Initial commit
This commit is contained in:
parent
fe51ac42df
commit
f00efdb5c4
48
README.md
48
README.md
|
|
@ -1,34 +1,40 @@
|
|||
# 基于scan context和CNN的重定位研究
|
||||
# scan context
|
||||
|
||||
#1-Day Learning, 1-Year Localization: Long-Term LiDAR Localization Using Scan Context Image
|
||||
# Scan Context: Egocentric Spatial Descriptor for Place Recognition within 3D Point Cloud Map
|
||||
|
||||
2019 ICRA
|
||||
**2018 IROS Giseop Kim and Ayoung Kim**
|
||||
|
||||
##SCI localization framework
|
||||
## Background
|
||||
|
||||

|
||||
- 回环检测(场景识别)=场景描述+搜索
|
||||
- 3D点云缺乏色彩信息,纹理信息等,无法提取出传统的图像所特有的特征(ORB,SIFT等)
|
||||
- 如果不对点云数据进行预处理的话,就只能进行几何匹配,消耗较高
|
||||
|
||||
##SCI
|
||||
## challenge
|
||||
|
||||
通过实验验证了与单通道图像训练相比的一点改 进。提出的sci比sc具有更高的分辨能力,是一种更适合cnn输入的格式。这个过程如图所示。我们注意到,进一步研究单色图像或彩色地图选择的网络调谐可以提高定位性能。
|
||||
- 降维的形式,尽可能多的保留深度信息
|
||||
- 描述符的编码
|
||||
- 相似度打分
|
||||
|
||||

|
||||
## Framework
|
||||
|
||||
Matlab使用colormap Jet 可以将灰度图像生成彩色的热度图,灰度值越高,色彩偏向暖色调。相反亦然。
|
||||
|
||||
|
||||

|
||||
## scan-context
|
||||
|
||||
##CNN选择
|
||||
cnn网络
|
||||

|
||||
输入:
|
||||
1.N-way SCI Augmentation
|
||||
2.热编码向量指示类别
|
||||
输出:
|
||||
scorevector
|
||||
|
||||

|
||||
将点云分为环形的一块一块,每一块的数值就是这一块点云海拔最高值。这样就实现了降维。
|
||||
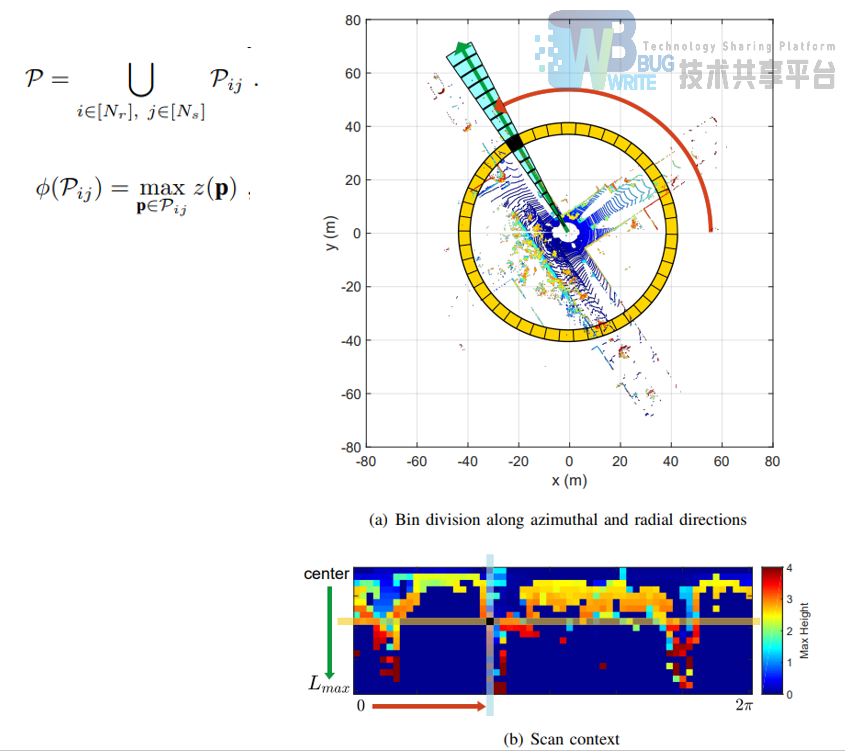
|
||||
|
||||
##Unseen place
|
||||
## Similarity Score between Scan Contexts
|
||||
|
||||

|
||||
由于雷达视角的不同,即当雷达在同一地点纯转动了一定角度之后,列向量向量值不变,但是会出现偏移;行向量的行为是向量中元素的顺序会发生改变,但是行向量不会发生偏移。采用列向比较。
|
||||
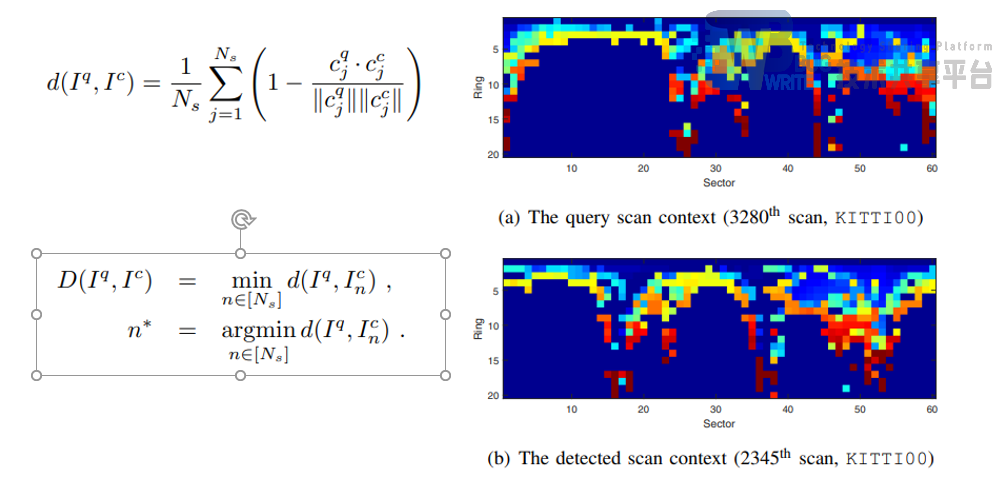
|
||||
|
||||
## Two-phase Search Algorithm
|
||||
|
||||
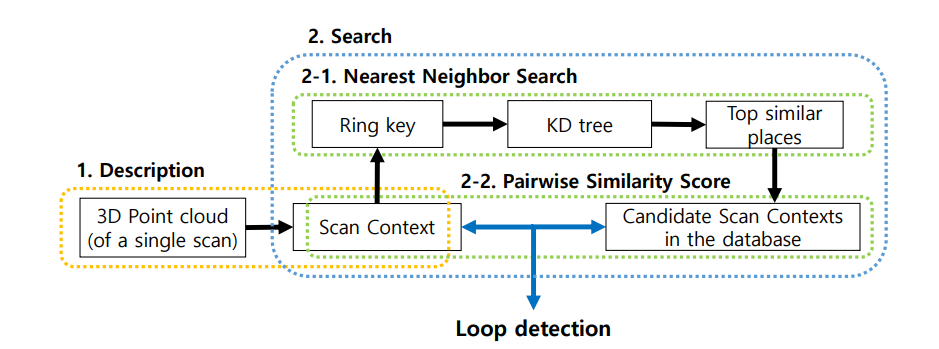
- 利用ring key 构造KD—Tree后最近邻检索
|
||||
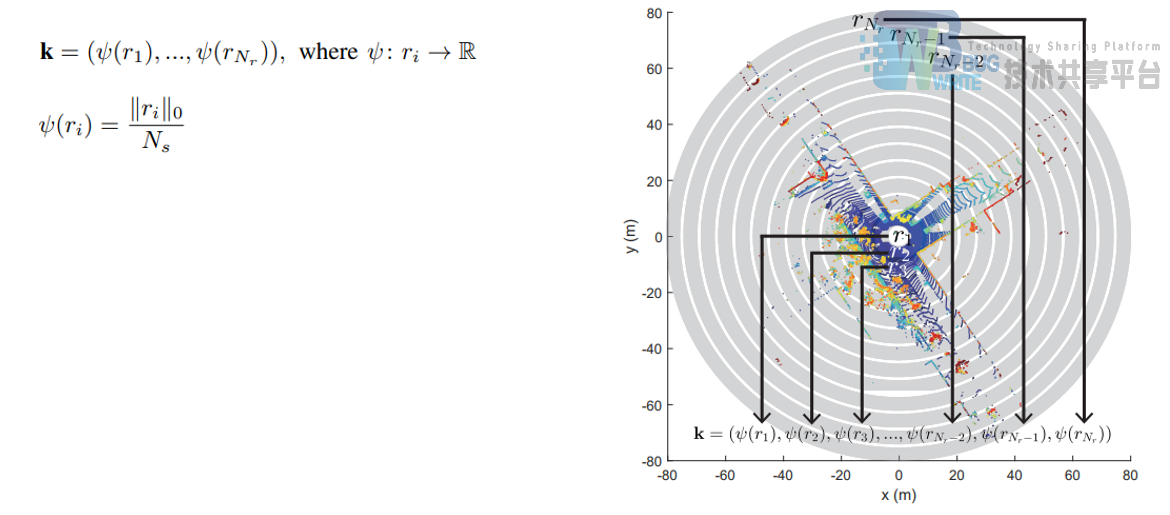
|
||||
- 相似度评分
|
||||
|
||||
- 找到闭环对应帧后使用ICP
|
||||
|
|
@ -1,92 +0,0 @@
|
|||
<!-- md preview: Show the rendered HTML markdown to the right of the current editor using ctrl-shift-m.-->
|
||||
|
||||
# Scan Context
|
||||
|
||||
## NEWS (Nov, 2020): integrated with LIO-SAM
|
||||
- A Scan Context integration for LIO-SAM, named [SC-LIO-SAM (link)](https://github.com/gisbi-kim/SC-LIO-SAM), is also released.
|
||||
|
||||
## NEWS (Oct, 2020): Radar Scan Context
|
||||
- An evaluation code for radar place recognition (a.k.a. Radar Scan Context) is uploaded.
|
||||
- please see the *fast_evaluator_radar* directory.
|
||||
|
||||
## NEWS (April, 2020): C++ implementation
|
||||
- C++ implementation released!
|
||||
- See the directory `cpp/module/Scancontext`
|
||||
- Features
|
||||
- Light-weight: a single header and cpp file named "Scancontext.h" and "Scancontext.cpp"
|
||||
- Our module has KDtree and we used <a href="https://github.com/jlblancoc/nanoflann"> nanoflann</a>. nanoflann is an also single-header-program and that file is in our directory.
|
||||
- Easy to use: A user just remembers and uses only two API functions; `makeAndSaveScancontextAndKeys` and `detectLoopClosureID`.
|
||||
- Fast: tested the loop detector runs at 10-15Hz (for 20 x 60 size, 10 candidates)
|
||||
- Example: Real-time LiDAR SLAM
|
||||
- We integrated the C++ implementation within the recent popular LiDAR odometry code, <a href="https://github.com/RobustFieldAutonomyLab/LeGO-LOAM"> LeGO-LOAM </a>.
|
||||
- That is, LiDAR SLAM = LiDAR Odometry (LeGO-LOAM) + Loop detection (Scan Context) and closure (GTSAM)
|
||||
- For details, see `cpp/example/lidar_slam` or refer this <a href="https://github.com/irapkaist/SC-LeGO-LOAM"> repository (SC-LeGO-LOAM)</a>.
|
||||
---
|
||||
|
||||
|
||||
- Scan Context is a global descriptor for LiDAR point cloud, which is proposed in this paper and details are easily summarized in this <a href="https://www.youtube.com/watch?v=_etNafgQXoY"> video </a>.
|
||||
|
||||
```
|
||||
@INPROCEEDINGS { gkim-2018-iros,
|
||||
author = {Kim, Giseop and Kim, Ayoung},
|
||||
title = { Scan Context: Egocentric Spatial Descriptor for Place Recognition within {3D} Point Cloud Map },
|
||||
booktitle = { Proceedings of the IEEE/RSJ International Conference on Intelligent Robots and Systems },
|
||||
year = { 2018 },
|
||||
month = { Oct. },
|
||||
address = { Madrid }
|
||||
}
|
||||
```
|
||||
- This point cloud descriptor is used for place retrieval problem such as place
|
||||
recognition and long-term localization.
|
||||
|
||||
|
||||
## What is Scan Context?
|
||||
|
||||
- Scan Context is a global descriptor for LiDAR point cloud, which is especially designed for a sparse and noisy point cloud acquired in outdoor environment.
|
||||
- It encodes egocentric visible information as below:
|
||||
<p align="center"><img src="example/basic/scmaking.gif" width=400></p>
|
||||
|
||||
- A user can vary the resolution of a Scan Context. Below is the example of Scan Contexts' various resolutions for the same point cloud.
|
||||
<p align="center"><img src="example/basic/various_res.png" width=300></p>
|
||||
|
||||
|
||||
## How to use?: example cases
|
||||
- The structure of this repository is composed of 3 example use cases.
|
||||
- Most of the codes are written in Matlab.
|
||||
- A directory _matlab_ contains main functions including Scan Context generation and the distance function.
|
||||
- A directory _example_ contains a full example code for a few applications. We provide a total 3 examples.
|
||||
1. _**basics**_ contains a literally basic codes such as generation and can be a start point to understand Scan Context.
|
||||
|
||||
2. _**place recognition**_ is an example directory for our IROS18 paper. The example is conducted using KITTI sequence 00 and PlaceRecognizer.m is the main code. You can easily grasp the full pipeline of Scan Context-based place recognition via watching and following the PlaceRecognizer.m code. Our Scan Context-based place recognition system consists of two steps; description and search. The search step is then composed of two hierarchical stages (1. ring key-based KD tree for fast candidate proposal, 2. candidate to query pairwise comparison-based nearest search). We note that our coarse yaw aligning-based pairwise distance enables reverse-revisit detection well, unlike others. The pipeline is below.
|
||||
<p align="center"><img src="example/place_recognition/sc_pipeline.png" width=600></p>
|
||||
|
||||
3. _**long-term localization**_ is an example directory for our RAL19 paper. For the separation of mapping and localization, there are separated train and test steps. The main training and test codes are written in python and Keras, only excluding data generation and performance evaluation codes (they are written in Matlab), and those python codes are provided using jupyter notebook. We note that some path may not directly work for your environment but the evaluation codes (e.g., makeDataForPRcurveForSCIresult.m) will help you understand how this classification-based SCI-localization system works. The figure below depicts our long-term localization pipeline. <p align="center"><img src="example/longterm_localization/sci_pipeline.png" width=600></p> More details of our long-term localization pipeline is found in the below paper and we also recommend you to watch this <a href="https://www.youtube.com/watch?v=apmmduXTnaE"> video </a>.
|
||||
```
|
||||
@ARTICLE{ gkim-2019-ral,
|
||||
author = {G. {Kim} and B. {Park} and A. {Kim}},
|
||||
journal = {IEEE Robotics and Automation Letters},
|
||||
title = {1-Day Learning, 1-Year Localization: Long-Term LiDAR Localization Using Scan Context Image},
|
||||
year = {2019},
|
||||
volume = {4},
|
||||
number = {2},
|
||||
pages = {1948-1955},
|
||||
month = {April}
|
||||
}
|
||||
```
|
||||
|
||||
4. _**SLAM**_ directory contains the practical use case of Scan Context for SLAM pipeline. The details are maintained in the related other repository _[PyICP SLAM](https://github.com/kissb2/PyICP-SLAM)_; the full-python LiDAR SLAM codes using Scan Context as a loop detector.
|
||||
|
||||
## Acknowledgment
|
||||
This work is supported by the Korea Agency for Infrastructure Technology Advancement (KAIA) grant funded by the Ministry of Land, Infrastructure and Transport of Korea (19CTAP-C142170-02), and [High-Definition Map Based Precise Vehicle Localization Using Cameras and LIDARs] project funded by NAVER LABS Corporation.
|
||||
|
||||
## Contact
|
||||
If you have any questions, contact here please
|
||||
```
|
||||
paulgkim@kaist.ac.kr
|
||||
```
|
||||
|
||||
## License
|
||||
<a rel="license" href="http://creativecommons.org/licenses/by-nc-sa/4.0/"><img alt="Creative Commons License" style="border-width:0" src="https://i.creativecommons.org/l/by-nc-sa/4.0/88x31.png" /></a><br />This work is licensed under a <a rel="license" href="http://creativecommons.org/licenses/by-nc-sa/4.0/">Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License</a>.
|
||||
|
||||
### Copyright
|
||||
- All codes on this page are copyrighted by KAIST and Naver Labs and published under the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License. You must attribute the work in the manner specified by the author. You may not use the work for commercial purposes, and you may only distribute the resulting work under the same license if you alter, transform, or create the work.
|
||||
|
|
@ -1,2 +0,0 @@
|
|||
# Go to
|
||||
- https://github.com/irapkaist/SC-LeGO-LOAM
|
||||
|
|
@ -1,117 +0,0 @@
|
|||
/***********************************************************************
|
||||
* Software License Agreement (BSD License)
|
||||
*
|
||||
* Copyright 2011-16 Jose Luis Blanco (joseluisblancoc@gmail.com).
|
||||
* All rights reserved.
|
||||
*
|
||||
* Redistribution and use in source and binary forms, with or without
|
||||
* modification, are permitted provided that the following conditions
|
||||
* are met:
|
||||
*
|
||||
* 1. Redistributions of source code must retain the above copyright
|
||||
* notice, this list of conditions and the following disclaimer.
|
||||
* 2. Redistributions in binary form must reproduce the above copyright
|
||||
* notice, this list of conditions and the following disclaimer in the
|
||||
* documentation and/or other materials provided with the distribution.
|
||||
*
|
||||
* THIS SOFTWARE IS PROVIDED BY THE AUTHOR ``AS IS'' AND ANY EXPRESS OR
|
||||
* IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES
|
||||
* OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED.
|
||||
* IN NO EVENT SHALL THE AUTHOR BE LIABLE FOR ANY DIRECT, INDIRECT,
|
||||
* INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT
|
||||
* NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE,
|
||||
* DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY
|
||||
* THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT
|
||||
* (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF
|
||||
* THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
||||
*************************************************************************/
|
||||
|
||||
#pragma once
|
||||
|
||||
#include <nanoflann.hpp>
|
||||
|
||||
#include <vector>
|
||||
|
||||
// ===== This example shows how to use nanoflann with these types of containers: =======
|
||||
//typedef std::vector<std::vector<double> > my_vector_of_vectors_t;
|
||||
//typedef std::vector<Eigen::VectorXd> my_vector_of_vectors_t; // This requires #include <Eigen/Dense>
|
||||
// =====================================================================================
|
||||
|
||||
|
||||
/** A simple vector-of-vectors adaptor for nanoflann, without duplicating the storage.
|
||||
* The i'th vector represents a point in the state space.
|
||||
*
|
||||
* \tparam DIM If set to >0, it specifies a compile-time fixed dimensionality for the points in the data set, allowing more compiler optimizations.
|
||||
* \tparam num_t The type of the point coordinates (typically, double or float).
|
||||
* \tparam Distance The distance metric to use: nanoflann::metric_L1, nanoflann::metric_L2, nanoflann::metric_L2_Simple, etc.
|
||||
* \tparam IndexType The type for indices in the KD-tree index (typically, size_t of int)
|
||||
*/
|
||||
template <class VectorOfVectorsType, typename num_t = double, int DIM = -1, class Distance = nanoflann::metric_L2, typename IndexType = size_t>
|
||||
struct KDTreeVectorOfVectorsAdaptor
|
||||
{
|
||||
typedef KDTreeVectorOfVectorsAdaptor<VectorOfVectorsType,num_t,DIM,Distance> self_t;
|
||||
typedef typename Distance::template traits<num_t,self_t>::distance_t metric_t;
|
||||
typedef nanoflann::KDTreeSingleIndexAdaptor< metric_t,self_t,DIM,IndexType> index_t;
|
||||
|
||||
index_t* index; //! The kd-tree index for the user to call its methods as usual with any other FLANN index.
|
||||
|
||||
/// Constructor: takes a const ref to the vector of vectors object with the data points
|
||||
KDTreeVectorOfVectorsAdaptor(const size_t /* dimensionality */, const VectorOfVectorsType &mat, const int leaf_max_size = 10) : m_data(mat)
|
||||
{
|
||||
assert(mat.size() != 0 && mat[0].size() != 0);
|
||||
const size_t dims = mat[0].size();
|
||||
if (DIM>0 && static_cast<int>(dims) != DIM)
|
||||
throw std::runtime_error("Data set dimensionality does not match the 'DIM' template argument");
|
||||
index = new index_t( static_cast<int>(dims), *this /* adaptor */, nanoflann::KDTreeSingleIndexAdaptorParams(leaf_max_size ) );
|
||||
index->buildIndex();
|
||||
}
|
||||
|
||||
~KDTreeVectorOfVectorsAdaptor() {
|
||||
delete index;
|
||||
}
|
||||
|
||||
const VectorOfVectorsType &m_data;
|
||||
|
||||
/** Query for the \a num_closest closest points to a given point (entered as query_point[0:dim-1]).
|
||||
* Note that this is a short-cut method for index->findNeighbors().
|
||||
* The user can also call index->... methods as desired.
|
||||
* \note nChecks_IGNORED is ignored but kept for compatibility with the original FLANN interface.
|
||||
*/
|
||||
inline void query(const num_t *query_point, const size_t num_closest, IndexType *out_indices, num_t *out_distances_sq, const int nChecks_IGNORED = 10) const
|
||||
{
|
||||
nanoflann::KNNResultSet<num_t,IndexType> resultSet(num_closest);
|
||||
resultSet.init(out_indices, out_distances_sq);
|
||||
index->findNeighbors(resultSet, query_point, nanoflann::SearchParams());
|
||||
}
|
||||
|
||||
/** @name Interface expected by KDTreeSingleIndexAdaptor
|
||||
* @{ */
|
||||
|
||||
const self_t & derived() const {
|
||||
return *this;
|
||||
}
|
||||
self_t & derived() {
|
||||
return *this;
|
||||
}
|
||||
|
||||
// Must return the number of data points
|
||||
inline size_t kdtree_get_point_count() const {
|
||||
return m_data.size();
|
||||
}
|
||||
|
||||
// Returns the dim'th component of the idx'th point in the class:
|
||||
inline num_t kdtree_get_pt(const size_t idx, const size_t dim) const {
|
||||
return m_data[idx][dim];
|
||||
}
|
||||
|
||||
// Optional bounding-box computation: return false to default to a standard bbox computation loop.
|
||||
// Return true if the BBOX was already computed by the class and returned in "bb" so it can be avoided to redo it again.
|
||||
// Look at bb.size() to find out the expected dimensionality (e.g. 2 or 3 for point clouds)
|
||||
template <class BBOX>
|
||||
bool kdtree_get_bbox(BBOX & /*bb*/) const {
|
||||
return false;
|
||||
}
|
||||
|
||||
/** @} */
|
||||
|
||||
}; // end of KDTreeVectorOfVectorsAdaptor
|
||||
|
|
@ -1,340 +0,0 @@
|
|||
#include "Scancontext.h"
|
||||
|
||||
// namespace SC2
|
||||
// {
|
||||
|
||||
void coreImportTest (void)
|
||||
{
|
||||
cout << "scancontext lib is successfully imported." << endl;
|
||||
} // coreImportTest
|
||||
|
||||
|
||||
float rad2deg(float radians)
|
||||
{
|
||||
return radians * 180.0 / M_PI;
|
||||
}
|
||||
|
||||
float deg2rad(float degrees)
|
||||
{
|
||||
return degrees * M_PI / 180.0;
|
||||
}
|
||||
|
||||
|
||||
float xy2theta( const float & _x, const float & _y )
|
||||
{
|
||||
if ( _x >= 0 & _y >= 0)
|
||||
return (180/M_PI) * atan(_y / _x);
|
||||
|
||||
if ( _x < 0 & _y >= 0)
|
||||
return 180 - ( (180/M_PI) * atan(_y / (-_x)) );
|
||||
|
||||
if ( _x < 0 & _y < 0)
|
||||
return 180 + ( (180/M_PI) * atan(_y / _x) );
|
||||
|
||||
if ( _x >= 0 & _y < 0)
|
||||
return 360 - ( (180/M_PI) * atan((-_y) / _x) );
|
||||
} // xy2theta
|
||||
|
||||
|
||||
MatrixXd circshift( MatrixXd &_mat, int _num_shift )
|
||||
{
|
||||
// shift columns to right direction
|
||||
assert(_num_shift >= 0);
|
||||
|
||||
if( _num_shift == 0 )
|
||||
{
|
||||
MatrixXd shifted_mat( _mat );
|
||||
return shifted_mat; // Early return
|
||||
}
|
||||
|
||||
MatrixXd shifted_mat = MatrixXd::Zero( _mat.rows(), _mat.cols() );
|
||||
for ( int col_idx = 0; col_idx < _mat.cols(); col_idx++ )
|
||||
{
|
||||
int new_location = (col_idx + _num_shift) % _mat.cols();
|
||||
shifted_mat.col(new_location) = _mat.col(col_idx);
|
||||
}
|
||||
|
||||
return shifted_mat;
|
||||
|
||||
} // circshift
|
||||
|
||||
|
||||
std::vector<float> eig2stdvec( MatrixXd _eigmat )
|
||||
{
|
||||
std::vector<float> vec( _eigmat.data(), _eigmat.data() + _eigmat.size() );
|
||||
return vec;
|
||||
} // eig2stdvec
|
||||
|
||||
|
||||
double SCManager::distDirectSC ( MatrixXd &_sc1, MatrixXd &_sc2 )
|
||||
{
|
||||
int num_eff_cols = 0; // i.e., to exclude all-nonzero sector
|
||||
double sum_sector_similarity = 0;
|
||||
for ( int col_idx = 0; col_idx < _sc1.cols(); col_idx++ )
|
||||
{
|
||||
VectorXd col_sc1 = _sc1.col(col_idx);
|
||||
VectorXd col_sc2 = _sc2.col(col_idx);
|
||||
|
||||
if( col_sc1.norm() == 0 | col_sc2.norm() == 0 )
|
||||
continue; // don't count this sector pair.
|
||||
|
||||
double sector_similarity = col_sc1.dot(col_sc2) / (col_sc1.norm() * col_sc2.norm());
|
||||
|
||||
sum_sector_similarity = sum_sector_similarity + sector_similarity;
|
||||
num_eff_cols = num_eff_cols + 1;
|
||||
}
|
||||
|
||||
double sc_sim = sum_sector_similarity / num_eff_cols;
|
||||
return 1.0 - sc_sim;
|
||||
|
||||
} // distDirectSC
|
||||
|
||||
|
||||
int SCManager::fastAlignUsingVkey( MatrixXd & _vkey1, MatrixXd & _vkey2)
|
||||
{
|
||||
int argmin_vkey_shift = 0;
|
||||
double min_veky_diff_norm = 10000000;
|
||||
for ( int shift_idx = 0; shift_idx < _vkey1.cols(); shift_idx++ )
|
||||
{
|
||||
MatrixXd vkey2_shifted = circshift(_vkey2, shift_idx);
|
||||
|
||||
MatrixXd vkey_diff = _vkey1 - vkey2_shifted;
|
||||
|
||||
double cur_diff_norm = vkey_diff.norm();
|
||||
if( cur_diff_norm < min_veky_diff_norm )
|
||||
{
|
||||
argmin_vkey_shift = shift_idx;
|
||||
min_veky_diff_norm = cur_diff_norm;
|
||||
}
|
||||
}
|
||||
|
||||
return argmin_vkey_shift;
|
||||
|
||||
} // fastAlignUsingVkey
|
||||
|
||||
|
||||
std::pair<double, int> SCManager::distanceBtnScanContext( MatrixXd &_sc1, MatrixXd &_sc2 )
|
||||
{
|
||||
// 1. fast align using variant key (not in original IROS18)
|
||||
MatrixXd vkey_sc1 = makeSectorkeyFromScancontext( _sc1 );
|
||||
MatrixXd vkey_sc2 = makeSectorkeyFromScancontext( _sc2 );
|
||||
int argmin_vkey_shift = fastAlignUsingVkey( vkey_sc1, vkey_sc2 );
|
||||
|
||||
const int SEARCH_RADIUS = round( 0.5 * SEARCH_RATIO * _sc1.cols() ); // a half of search range
|
||||
std::vector<int> shift_idx_search_space { argmin_vkey_shift };
|
||||
for ( int ii = 1; ii < SEARCH_RADIUS + 1; ii++ )
|
||||
{
|
||||
shift_idx_search_space.push_back( (argmin_vkey_shift + ii + _sc1.cols()) % _sc1.cols() );
|
||||
shift_idx_search_space.push_back( (argmin_vkey_shift - ii + _sc1.cols()) % _sc1.cols() );
|
||||
}
|
||||
std::sort(shift_idx_search_space.begin(), shift_idx_search_space.end());
|
||||
|
||||
// 2. fast columnwise diff
|
||||
int argmin_shift = 0;
|
||||
double min_sc_dist = 10000000;
|
||||
for ( int num_shift: shift_idx_search_space )
|
||||
{
|
||||
MatrixXd sc2_shifted = circshift(_sc2, num_shift);
|
||||
double cur_sc_dist = distDirectSC( _sc1, sc2_shifted );
|
||||
if( cur_sc_dist < min_sc_dist )
|
||||
{
|
||||
argmin_shift = num_shift;
|
||||
min_sc_dist = cur_sc_dist;
|
||||
}
|
||||
}
|
||||
|
||||
return make_pair(min_sc_dist, argmin_shift);
|
||||
|
||||
} // distanceBtnScanContext
|
||||
|
||||
|
||||
MatrixXd SCManager::makeScancontext( pcl::PointCloud<SCPointType> & _scan_down )
|
||||
{
|
||||
TicToc t_making_desc;
|
||||
|
||||
int num_pts_scan_down = _scan_down.points.size();
|
||||
|
||||
// main
|
||||
const int NO_POINT = -1000;
|
||||
MatrixXd desc = NO_POINT * MatrixXd::Ones(PC_NUM_RING, PC_NUM_SECTOR);
|
||||
|
||||
SCPointType pt;
|
||||
float azim_angle, azim_range; // wihtin 2d plane
|
||||
int ring_idx, sctor_idx;
|
||||
for (int pt_idx = 0; pt_idx < num_pts_scan_down; pt_idx++)
|
||||
{
|
||||
pt.x = _scan_down.points[pt_idx].x;
|
||||
pt.y = _scan_down.points[pt_idx].y;
|
||||
pt.z = _scan_down.points[pt_idx].z + LIDAR_HEIGHT; // naive adding is ok (all points should be > 0).
|
||||
|
||||
// xyz to ring, sector
|
||||
azim_range = sqrt(pt.x * pt.x + pt.y * pt.y);
|
||||
azim_angle = xy2theta(pt.x, pt.y);
|
||||
|
||||
// if range is out of roi, pass
|
||||
if( azim_range > PC_MAX_RADIUS )
|
||||
continue;
|
||||
|
||||
ring_idx = std::max( std::min( PC_NUM_RING, int(ceil( (azim_range / PC_MAX_RADIUS) * PC_NUM_RING )) ), 1 );
|
||||
sctor_idx = std::max( std::min( PC_NUM_SECTOR, int(ceil( (azim_angle / 360.0) * PC_NUM_SECTOR )) ), 1 );
|
||||
|
||||
// taking maximum z
|
||||
if ( desc(ring_idx-1, sctor_idx-1) < pt.z ) // -1 means cpp starts from 0
|
||||
desc(ring_idx-1, sctor_idx-1) = pt.z; // update for taking maximum value at that bin
|
||||
}
|
||||
|
||||
// reset no points to zero (for cosine dist later)
|
||||
for ( int row_idx = 0; row_idx < desc.rows(); row_idx++ )
|
||||
for ( int col_idx = 0; col_idx < desc.cols(); col_idx++ )
|
||||
if( desc(row_idx, col_idx) == NO_POINT )
|
||||
desc(row_idx, col_idx) = 0;
|
||||
|
||||
t_making_desc.toc("PolarContext making");
|
||||
|
||||
return desc;
|
||||
} // SCManager::makeScancontext
|
||||
|
||||
|
||||
MatrixXd SCManager::makeRingkeyFromScancontext( Eigen::MatrixXd &_desc )
|
||||
{
|
||||
/*
|
||||
* summary: rowwise mean vector
|
||||
*/
|
||||
Eigen::MatrixXd invariant_key(_desc.rows(), 1);
|
||||
for ( int row_idx = 0; row_idx < _desc.rows(); row_idx++ )
|
||||
{
|
||||
Eigen::MatrixXd curr_row = _desc.row(row_idx);
|
||||
invariant_key(row_idx, 0) = curr_row.mean();
|
||||
}
|
||||
|
||||
return invariant_key;
|
||||
} // SCManager::makeRingkeyFromScancontext
|
||||
|
||||
|
||||
MatrixXd SCManager::makeSectorkeyFromScancontext( Eigen::MatrixXd &_desc )
|
||||
{
|
||||
/*
|
||||
* summary: columnwise mean vector
|
||||
*/
|
||||
Eigen::MatrixXd variant_key(1, _desc.cols());
|
||||
for ( int col_idx = 0; col_idx < _desc.cols(); col_idx++ )
|
||||
{
|
||||
Eigen::MatrixXd curr_col = _desc.col(col_idx);
|
||||
variant_key(0, col_idx) = curr_col.mean();
|
||||
}
|
||||
|
||||
return variant_key;
|
||||
} // SCManager::makeSectorkeyFromScancontext
|
||||
|
||||
|
||||
void SCManager::makeAndSaveScancontextAndKeys( pcl::PointCloud<SCPointType> & _scan_down )
|
||||
{
|
||||
Eigen::MatrixXd sc = makeScancontext(_scan_down); // v1
|
||||
Eigen::MatrixXd ringkey = makeRingkeyFromScancontext( sc );
|
||||
Eigen::MatrixXd sectorkey = makeSectorkeyFromScancontext( sc );
|
||||
std::vector<float> polarcontext_invkey_vec = eig2stdvec( ringkey );
|
||||
|
||||
polarcontexts_.push_back( sc );
|
||||
polarcontext_invkeys_.push_back( ringkey );
|
||||
polarcontext_vkeys_.push_back( sectorkey );
|
||||
polarcontext_invkeys_mat_.push_back( polarcontext_invkey_vec );
|
||||
|
||||
// cout <<polarcontext_vkeys_.size() << endl;
|
||||
|
||||
} // SCManager::makeAndSaveScancontextAndKeys
|
||||
|
||||
|
||||
std::pair<int, float> SCManager::detectLoopClosureID ( void )
|
||||
{
|
||||
int loop_id { -1 }; // init with -1, -1 means no loop (== LeGO-LOAM's variable "closestHistoryFrameID")
|
||||
|
||||
auto curr_key = polarcontext_invkeys_mat_.back(); // current observation (query)
|
||||
auto curr_desc = polarcontexts_.back(); // current observation (query)
|
||||
|
||||
/*
|
||||
* step 1: candidates from ringkey tree_
|
||||
*/
|
||||
if( polarcontext_invkeys_mat_.size() < NUM_EXCLUDE_RECENT + 1)
|
||||
{
|
||||
std::pair<int, float> result {loop_id, 0.0};
|
||||
return result; // Early return
|
||||
}
|
||||
|
||||
// tree_ reconstruction (not mandatory to make everytime)
|
||||
if( tree_making_period_conter % TREE_MAKING_PERIOD_ == 0) // to save computation cost
|
||||
{
|
||||
TicToc t_tree_construction;
|
||||
|
||||
polarcontext_invkeys_to_search_.clear();
|
||||
polarcontext_invkeys_to_search_.assign( polarcontext_invkeys_mat_.begin(), polarcontext_invkeys_mat_.end() - NUM_EXCLUDE_RECENT ) ;
|
||||
|
||||
polarcontext_tree_.reset();
|
||||
polarcontext_tree_ = std::make_unique<InvKeyTree>(PC_NUM_RING /* dim */, polarcontext_invkeys_to_search_, 10 /* max leaf */ );
|
||||
// tree_ptr_->index->buildIndex(); // inernally called in the constructor of InvKeyTree (for detail, refer the nanoflann and KDtreeVectorOfVectorsAdaptor)
|
||||
t_tree_construction.toc("Tree construction");
|
||||
}
|
||||
tree_making_period_conter = tree_making_period_conter + 1;
|
||||
|
||||
double min_dist = 10000000; // init with somthing large
|
||||
int nn_align = 0;
|
||||
int nn_idx = 0;
|
||||
|
||||
// knn search
|
||||
std::vector<size_t> candidate_indexes( NUM_CANDIDATES_FROM_TREE );
|
||||
std::vector<float> out_dists_sqr( NUM_CANDIDATES_FROM_TREE );
|
||||
|
||||
TicToc t_tree_search;
|
||||
nanoflann::KNNResultSet<float> knnsearch_result( NUM_CANDIDATES_FROM_TREE );
|
||||
knnsearch_result.init( &candidate_indexes[0], &out_dists_sqr[0] );
|
||||
polarcontext_tree_->index->findNeighbors( knnsearch_result, &curr_key[0] /* query */, nanoflann::SearchParams(10) );
|
||||
t_tree_search.toc("Tree search");
|
||||
|
||||
/*
|
||||
* step 2: pairwise distance (find optimal columnwise best-fit using cosine distance)
|
||||
*/
|
||||
TicToc t_calc_dist;
|
||||
for ( int candidate_iter_idx = 0; candidate_iter_idx < NUM_CANDIDATES_FROM_TREE; candidate_iter_idx++ )
|
||||
{
|
||||
MatrixXd polarcontext_candidate = polarcontexts_[ candidate_indexes[candidate_iter_idx] ];
|
||||
std::pair<double, int> sc_dist_result = distanceBtnScanContext( curr_desc, polarcontext_candidate );
|
||||
|
||||
double candidate_dist = sc_dist_result.first;
|
||||
int candidate_align = sc_dist_result.second;
|
||||
|
||||
if( candidate_dist < min_dist )
|
||||
{
|
||||
min_dist = candidate_dist;
|
||||
nn_align = candidate_align;
|
||||
|
||||
nn_idx = candidate_indexes[candidate_iter_idx];
|
||||
}
|
||||
}
|
||||
t_calc_dist.toc("Distance calc");
|
||||
|
||||
/*
|
||||
* loop threshold check
|
||||
*/
|
||||
if( min_dist < SC_DIST_THRES )
|
||||
{
|
||||
loop_id = nn_idx;
|
||||
|
||||
// std::cout.precision(3);
|
||||
cout << "[Loop found] Nearest distance: " << min_dist << " btn " << polarcontexts_.size()-1 << " and " << nn_idx << "." << endl;
|
||||
cout << "[Loop found] yaw diff: " << nn_align * PC_UNIT_SECTORANGLE << " deg." << endl;
|
||||
}
|
||||
else
|
||||
{
|
||||
std::cout.precision(3);
|
||||
cout << "[Not loop] Nearest distance: " << min_dist << " btn " << polarcontexts_.size()-1 << " and " << nn_idx << "." << endl;
|
||||
cout << "[Not loop] yaw diff: " << nn_align * PC_UNIT_SECTORANGLE << " deg." << endl;
|
||||
}
|
||||
|
||||
// To do: return also nn_align (i.e., yaw diff)
|
||||
float yaw_diff_rad = deg2rad(nn_align * PC_UNIT_SECTORANGLE);
|
||||
std::pair<int, float> result {loop_id, yaw_diff_rad};
|
||||
|
||||
return result;
|
||||
|
||||
} // SCManager::detectLoopClosureID
|
||||
|
||||
// } // namespace SC2
|
||||
|
|
@ -1,110 +0,0 @@
|
|||
#pragma once
|
||||
|
||||
#include <ctime>
|
||||
#include <cassert>
|
||||
#include <cmath>
|
||||
#include <utility>
|
||||
#include <vector>
|
||||
#include <algorithm>
|
||||
#include <cstdlib>
|
||||
#include <memory>
|
||||
#include <iostream>
|
||||
|
||||
#include <Eigen/Dense>
|
||||
|
||||
#include <opencv2/opencv.hpp>
|
||||
#include <opencv2/core/eigen.hpp>
|
||||
#include <opencv2/highgui/highgui.hpp>
|
||||
#include <cv_bridge/cv_bridge.h>
|
||||
|
||||
#include <pcl/point_cloud.h>
|
||||
#include <pcl/point_types.h>
|
||||
#include <pcl/filters/voxel_grid.h>
|
||||
#include <pcl_conversions/pcl_conversions.h>
|
||||
|
||||
#include "nanoflann.hpp"
|
||||
#include "KDTreeVectorOfVectorsAdaptor.h"
|
||||
|
||||
#include "tictoc.h"
|
||||
|
||||
using namespace Eigen;
|
||||
using namespace nanoflann;
|
||||
|
||||
using std::cout;
|
||||
using std::endl;
|
||||
using std::make_pair;
|
||||
|
||||
using std::atan2;
|
||||
using std::cos;
|
||||
using std::sin;
|
||||
|
||||
using SCPointType = pcl::PointXYZI; // using xyz only. but a user can exchange the original bin encoding function (i.e., max hegiht) to max intensity (for detail, refer 20 ICRA Intensity Scan Context)
|
||||
using KeyMat = std::vector<std::vector<float> >;
|
||||
using InvKeyTree = KDTreeVectorOfVectorsAdaptor< KeyMat, float >;
|
||||
|
||||
|
||||
// namespace SC2
|
||||
// {
|
||||
|
||||
void coreImportTest ( void );
|
||||
|
||||
|
||||
// sc param-independent helper functions
|
||||
float xy2theta( const float & _x, const float & _y );
|
||||
MatrixXd circshift( MatrixXd &_mat, int _num_shift );
|
||||
std::vector<float> eig2stdvec( MatrixXd _eigmat );
|
||||
|
||||
|
||||
class SCManager
|
||||
{
|
||||
public:
|
||||
SCManager( ) = default; // reserving data space (of std::vector) could be considered. but the descriptor is lightweight so don't care.
|
||||
|
||||
Eigen::MatrixXd makeScancontext( pcl::PointCloud<SCPointType> & _scan_down );
|
||||
Eigen::MatrixXd makeRingkeyFromScancontext( Eigen::MatrixXd &_desc );
|
||||
Eigen::MatrixXd makeSectorkeyFromScancontext( Eigen::MatrixXd &_desc );
|
||||
|
||||
int fastAlignUsingVkey ( MatrixXd & _vkey1, MatrixXd & _vkey2 );
|
||||
double distDirectSC ( MatrixXd &_sc1, MatrixXd &_sc2 ); // "d" (eq 5) in the original paper (IROS 18)
|
||||
std::pair<double, int> distanceBtnScanContext ( MatrixXd &_sc1, MatrixXd &_sc2 ); // "D" (eq 6) in the original paper (IROS 18)
|
||||
|
||||
// User-side API
|
||||
void makeAndSaveScancontextAndKeys( pcl::PointCloud<SCPointType> & _scan_down );
|
||||
std::pair<int, float> detectLoopClosureID( void ); // int: nearest node index, float: relative yaw
|
||||
|
||||
public:
|
||||
// hyper parameters ()
|
||||
const double LIDAR_HEIGHT = 2.0; // lidar height : add this for simply directly using lidar scan in the lidar local coord (not robot base coord) / if you use robot-coord-transformed lidar scans, just set this as 0.
|
||||
|
||||
const int PC_NUM_RING = 20; // 20 in the original paper (IROS 18)
|
||||
const int PC_NUM_SECTOR = 60; // 60 in the original paper (IROS 18)
|
||||
const double PC_MAX_RADIUS = 80.0; // 80 meter max in the original paper (IROS 18)
|
||||
const double PC_UNIT_SECTORANGLE = 360.0 / double(PC_NUM_SECTOR);
|
||||
const double PC_UNIT_RINGGAP = PC_MAX_RADIUS / double(PC_NUM_RING);
|
||||
|
||||
// tree
|
||||
const int NUM_EXCLUDE_RECENT = 50; // simply just keyframe gap, but node position distance-based exclusion is ok.
|
||||
const int NUM_CANDIDATES_FROM_TREE = 10; // 10 is enough. (refer the IROS 18 paper)
|
||||
|
||||
// loop thres
|
||||
const double SEARCH_RATIO = 0.1; // for fast comparison, no Brute-force, but search 10 % is okay. // not was in the original conf paper, but improved ver.
|
||||
const double SC_DIST_THRES = 0.13; // empirically 0.1-0.2 is fine (rare false-alarms) for 20x60 polar context (but for 0.15 <, DCS or ICP fit score check (e.g., in LeGO-LOAM) should be required for robustness)
|
||||
// const double SC_DIST_THRES = 0.5; // 0.4-0.6 is good choice for using with robust kernel (e.g., Cauchy, DCS) + icp fitness threshold / if not, recommend 0.1-0.15
|
||||
|
||||
// config
|
||||
const int TREE_MAKING_PERIOD_ = 50; // i.e., remaking tree frequency, to avoid non-mandatory every remaking, to save time cost / if you want to find a very recent revisits use small value of it (it is enough fast ~ 5-50ms wrt N.).
|
||||
int tree_making_period_conter = 0;
|
||||
|
||||
// data
|
||||
std::vector<double> polarcontexts_timestamp_; // optional.
|
||||
std::vector<Eigen::MatrixXd> polarcontexts_;
|
||||
std::vector<Eigen::MatrixXd> polarcontext_invkeys_;
|
||||
std::vector<Eigen::MatrixXd> polarcontext_vkeys_;
|
||||
|
||||
KeyMat polarcontext_invkeys_mat_;
|
||||
KeyMat polarcontext_invkeys_to_search_;
|
||||
std::unique_ptr<InvKeyTree> polarcontext_tree_;
|
||||
|
||||
}; // SCManager
|
||||
|
||||
// } // namespace SC2
|
||||
File diff suppressed because it is too large
Load Diff
|
|
@ -1,47 +0,0 @@
|
|||
// Author: Tong Qin qintonguav@gmail.com
|
||||
// Shaozu Cao saozu.cao@connect.ust.hk
|
||||
|
||||
#pragma once
|
||||
|
||||
#include <ctime>
|
||||
#include <iostream>
|
||||
#include <string>
|
||||
#include <cstdlib>
|
||||
#include <chrono>
|
||||
|
||||
class TicToc
|
||||
{
|
||||
public:
|
||||
TicToc()
|
||||
{
|
||||
tic();
|
||||
}
|
||||
|
||||
TicToc( bool _disp )
|
||||
{
|
||||
disp_ = _disp;
|
||||
tic();
|
||||
}
|
||||
|
||||
void tic()
|
||||
{
|
||||
start = std::chrono::system_clock::now();
|
||||
}
|
||||
|
||||
void toc( std::string _about_task )
|
||||
{
|
||||
end = std::chrono::system_clock::now();
|
||||
std::chrono::duration<double> elapsed_seconds = end - start;
|
||||
double elapsed_ms = elapsed_seconds.count() * 1000;
|
||||
|
||||
if( disp_ )
|
||||
{
|
||||
std::cout.precision(3); // 10 for sec, 3 for ms
|
||||
std::cout << _about_task << ": " << elapsed_ms << " msec." << std::endl;
|
||||
}
|
||||
}
|
||||
|
||||
private:
|
||||
std::chrono::time_point<std::chrono::system_clock> start, end;
|
||||
bool disp_ = false;
|
||||
};
|
||||
|
|
@ -1,2 +0,0 @@
|
|||
# Scan Context for LiDAR SLAM
|
||||
- Go to [PyICP SLAM](https://github.com/kissb2/PyICP-SLAM)
|
||||
|
|
@ -1,81 +0,0 @@
|
|||
clear; clc;
|
||||
addpath(genpath('../../matlab/'));
|
||||
|
||||
|
||||
%% Parameters
|
||||
data_dir = '../../sample_data/KITTI/00/velodyne/';
|
||||
|
||||
basic_max_range = 80; % meter
|
||||
basic_num_sectors = 60;
|
||||
basic_num_rings = 20;
|
||||
|
||||
|
||||
%% Visualization of ScanContext
|
||||
bin_path = [data_dir, '000094.bin'];
|
||||
ptcloud = KITTIbin2Ptcloud(bin_path);
|
||||
sc = Ptcloud2ScanContext(ptcloud, basic_num_sectors, basic_num_rings, basic_max_range);
|
||||
|
||||
h1 = figure(1); clf;
|
||||
imagesc(sc);
|
||||
set(gcf, 'Position', [10 10 800 300]);
|
||||
xlabel('sector'); ylabel('ring');
|
||||
|
||||
% for vivid visualization
|
||||
colormap jet;
|
||||
caxis([0, 4]); % KITTI00 is usually in z: [0, 4]
|
||||
|
||||
|
||||
%% Making Ringkey and maintaining kd-tree
|
||||
% Read the PlaceRecognizer.m
|
||||
|
||||
|
||||
%% ScanContext with different resolution
|
||||
|
||||
figure(2); clf;
|
||||
pcshow(ptcloud); colormap jet; caxis([0 4]);
|
||||
|
||||
res = [0.25, 0.5, 1, 2, 3];
|
||||
|
||||
h2=figure(3); clf;
|
||||
set(gcf, 'Position', [10 10 500 1000]);
|
||||
|
||||
for i = 1:length(res)
|
||||
num_sectors = basic_num_sectors * res(i);
|
||||
num_rings = basic_num_rings * res(i);
|
||||
|
||||
sc = Ptcloud2ScanContext(ptcloud, num_sectors, num_rings, basic_max_range);
|
||||
|
||||
subplot(length(res), 1, i);
|
||||
imagesc(sc); hold on;
|
||||
colormap jet;
|
||||
caxis([0, 4]); % KITTI00 is usually in z: [0, 4]
|
||||
end
|
||||
|
||||
|
||||
|
||||
%% Comparison btn two scan contexts
|
||||
|
||||
KITTI_bin1a_path = [data_dir, '000094.bin'];
|
||||
KITTI_bin1b_path = [data_dir, '000095.bin'];
|
||||
KITTI_bin2a_path = [data_dir, '000198.bin'];
|
||||
KITTI_bin2b_path = [data_dir, '000199.bin'];
|
||||
|
||||
ptcloud_KITTI1a = KITTIbin2Ptcloud(KITTI_bin1a_path);
|
||||
ptcloud_KITTI1b = KITTIbin2Ptcloud(KITTI_bin1b_path);
|
||||
ptcloud_KITTI2a = KITTIbin2Ptcloud(KITTI_bin2a_path);
|
||||
ptcloud_KITTI2b = KITTIbin2Ptcloud(KITTI_bin2b_path);
|
||||
|
||||
sc_KITTI1a = Ptcloud2ScanContext(ptcloud_KITTI1a, basic_num_sectors, basic_num_rings, basic_max_range);
|
||||
sc_KITTI1b = Ptcloud2ScanContext(ptcloud_KITTI1b, basic_num_sectors, basic_num_rings, basic_max_range);
|
||||
sc_KITTI2a = Ptcloud2ScanContext(ptcloud_KITTI2a, basic_num_sectors, basic_num_rings, basic_max_range);
|
||||
sc_KITTI2b = Ptcloud2ScanContext(ptcloud_KITTI2b, basic_num_sectors, basic_num_rings, basic_max_range);
|
||||
|
||||
dist_1a_1b = DistanceBtnScanContexts(sc_KITTI1a, sc_KITTI1b);
|
||||
dist_1b_2a = DistanceBtnScanContexts(sc_KITTI1b, sc_KITTI2a);
|
||||
dist_1a_2a = DistanceBtnScanContexts(sc_KITTI1a, sc_KITTI2a);
|
||||
dist_2a_2b = DistanceBtnScanContexts(sc_KITTI2a, sc_KITTI2b);
|
||||
|
||||
disp([dist_1a_1b, dist_1b_2a, dist_1a_2a, dist_2a_2b]);
|
||||
|
||||
|
||||
|
||||
Binary file not shown.
|
Before Width: | Height: | Size: 108 KiB |
Binary file not shown.
|
Before Width: | Height: | Size: 2.0 MiB |
Binary file not shown.
|
Before Width: | Height: | Size: 25 KiB |
|
|
@ -1,112 +0,0 @@
|
|||
%% information
|
||||
% main for Sampling places
|
||||
|
||||
%%
|
||||
clear; clc;
|
||||
addpath(genpath('../../../../../matlab/'));
|
||||
addpath(genpath('./helper'));
|
||||
|
||||
SaveDirectoryList
|
||||
Parameters
|
||||
|
||||
%% Preparation 1: make pre-determined Grid Cell index
|
||||
PlaceIndexAndGridCenters_10m = makeGridCellIndex(xRange, yRange, 10);
|
||||
|
||||
%% Preparation 2: get scan times
|
||||
SequenceDate = '2012-01-15'; % ### Change this part to your date
|
||||
ScanBaseDir = 'F:\NCLT/'; % ### Change this part to your path
|
||||
|
||||
ScanDir = strcat(ScanBaseDir, SequenceDate, '/velodyne_sync/');
|
||||
Scans = dir(ScanDir); Scans(1:2, :) = []; Scans = {Scans(:).name};
|
||||
ScanTimes = getNCLTscanInformation(Scans);
|
||||
|
||||
%% Preparation 3: load GT pose (for calc moving diff and location)
|
||||
GroundTruthPosePath = strcat(ScanBaseDir, SequenceDate, '/groundtruth_', SequenceDate, '.csv');
|
||||
GroundTruthPoseData = csvread(GroundTruthPosePath);
|
||||
|
||||
GroundTruthPoseTime = GroundTruthPoseData(:, 1);
|
||||
GroundTruthPoseXYZ = GroundTruthPoseData(:, 2:4);
|
||||
|
||||
nGroundTruthPoses = length(GroundTruthPoseData);
|
||||
|
||||
%% logger
|
||||
TrajectoryInformationWRT10mCell = [];
|
||||
|
||||
nTotalSampledPlaces = 0;
|
||||
|
||||
%% Main: Sampling
|
||||
|
||||
MoveCounter = 0; % Reset 0 again for every SamplingGap reached.
|
||||
for ii = 1000:nGroundTruthPoses % just quite large number 1000 for avoiding first several NaNs
|
||||
curTime = GroundTruthPoseTime(ii, 1);
|
||||
|
||||
prvPose = GroundTruthPoseXYZ(ii-1, :);
|
||||
curPose = GroundTruthPoseXYZ(ii, :);
|
||||
|
||||
curMove = norm(curPose - prvPose);
|
||||
MoveCounter = MoveCounter + curMove;
|
||||
|
||||
if(MoveCounter >= SamplingGap)
|
||||
nTotalSampledPlaces = nTotalSampledPlaces + 1;
|
||||
curSamplingCounter = nTotalSampledPlaces;
|
||||
|
||||
% Returns the index of the cell, where the current pose is closest to the cell's center coordinates.
|
||||
PlaceIdx_10m = getPlaceIdx(curPose, PlaceIndexAndGridCenters_10m); % 2nd argument is cell's size
|
||||
|
||||
% load current point cloud
|
||||
curPtcloud = getNearestPtcloud( ScanTimes, curTime, Scans, ScanDir);
|
||||
|
||||
%% Save data
|
||||
% log
|
||||
TrajectoryInformationWRT10mCell = [TrajectoryInformationWRT10mCell; curTime, curPose, nTotalSampledPlaces, PlaceIdx_10m];
|
||||
|
||||
% scan context
|
||||
ScanContextForward = Ptcloud2ScanContext(curPtcloud, nSectors, nRings, Lmax);
|
||||
|
||||
% SCI gray (1 channel)
|
||||
ScanContextForwardRanged = ScaleSc2Img(ScanContextForward, NCLTminHeight, NCLTmaxHeight);
|
||||
ScanContextForwardScaled = ScanContextForwardRanged./maxColor;
|
||||
|
||||
SCIforwardGray = round(ScanContextForwardScaled*255);
|
||||
SCIforwardGray = ind2gray(SCIforwardGray, gray(255));
|
||||
|
||||
% SCI jet (color, 3 channel)
|
||||
SCIforwardColor = round(ScanContextForwardScaled*255);
|
||||
SCIforwardColor = ind2rgb(SCIforwardColor, jet(255));
|
||||
|
||||
saveSCIcolor(SCIforwardColor, DIR_SCIcolor, curSamplingCounter, PlaceIdx_10m, '10', 'f');
|
||||
|
||||
% SCI jet + Backward dataAug
|
||||
ScanContextBackwardScaled = circshift(ScanContextForwardScaled, nSectors/2, 2);
|
||||
SCIbackwardColor = round(ScanContextBackwardScaled*255);
|
||||
SCIbackwardColor = ind2rgb(SCIbackwardColor, jet(255));
|
||||
|
||||
saveSCIcolor(SCIforwardColor, DIR_SCIcolorAlsoBack, curSamplingCounter, PlaceIdx_10m, '10', 'f');
|
||||
saveSCIcolor(SCIbackwardColor, DIR_SCIcolorAlsoBack, curSamplingCounter, PlaceIdx_10m, '10', 'b');
|
||||
|
||||
% End: Reset counter
|
||||
MoveCounter = 0;
|
||||
|
||||
% Tracking progress message
|
||||
if(rem(curSamplingCounter, 100) == 0)
|
||||
message = strcat(num2str(curSamplingCounter), "th sample is saved." );
|
||||
disp(message)
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
||||
%% save Trajectory Information
|
||||
% 10m
|
||||
filepath = strcat(DIR_SampledPlacesInformation, '/TrajectoryInformation.csv');
|
||||
TrajectoryInformation = TrajectoryInformationWRT10mCell;
|
||||
dlmwrite(filepath, TrajectoryInformation, 'precision','%.6f')
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
|
@ -1,29 +0,0 @@
|
|||
% flag for train/test
|
||||
IF_TRAINING = 1;
|
||||
|
||||
% sampling gap
|
||||
SamplingGap = 1; % in meter
|
||||
|
||||
% place resolution
|
||||
PlaceCellSize = 10; % in meter
|
||||
|
||||
% NCLT region
|
||||
xRange = [-350, 130];
|
||||
yRange = [-730, 120];
|
||||
|
||||
|
||||
% scan context
|
||||
nRings = 40;
|
||||
nSectors = 120;
|
||||
Lmax = 80;
|
||||
|
||||
% scan context image
|
||||
NCLTminHeight = 0;
|
||||
NCLTmaxHeight = 15;
|
||||
|
||||
SCI_HEIGHT_RANGE = [NCLTminHeight, NCLTmaxHeight];
|
||||
|
||||
minColor = 0;
|
||||
maxColor = 255;
|
||||
rangeColor = maxColor - minColor;
|
||||
|
||||
|
|
@ -1,6 +0,0 @@
|
|||
|
||||
%% Save Directories
|
||||
|
||||
DIR_SampledPlacesInformation = './data/SampledPlacesInformation/';
|
||||
DIR_SCIcolor = './data/SCI_jet0to15/';
|
||||
DIR_SCIcolorAlsoBack = './data/SCI_jet0to15_BackAug/';
|
||||
|
|
@ -1,33 +0,0 @@
|
|||
function [ scScaled ] = ScaleSc2Img( scOriginal, minHeight, maxHeight )
|
||||
|
||||
maxColor = 255;
|
||||
|
||||
rangeHeight = maxHeight - minHeight;
|
||||
|
||||
nRows = size(scOriginal, 1);
|
||||
nCols = size(scOriginal, 2);
|
||||
|
||||
scOriginalRangeCut = scOriginal;
|
||||
% cut into range
|
||||
for ithRow = 1:nRows
|
||||
for jthCol = 1:nCols
|
||||
|
||||
ithPixel = scOriginal(ithRow, jthCol);
|
||||
|
||||
if(ithPixel >= maxHeight)
|
||||
scOriginalRangeCut(ithRow, jthCol) = maxHeight;
|
||||
end
|
||||
|
||||
if(ithPixel <= minHeight)
|
||||
scOriginalRangeCut(ithRow, jthCol) = minHeight;
|
||||
end
|
||||
|
||||
scOriginalRangeCut(ithRow, jthCol) = round(scOriginalRangeCut(ithRow, jthCol) * (maxColor/rangeHeight));
|
||||
end
|
||||
end
|
||||
|
||||
scScaled = scOriginalRangeCut;
|
||||
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,15 +0,0 @@
|
|||
function [ ScanTimes ] = getNCLTscanInformation( Scans )
|
||||
|
||||
nScans = length(Scans);
|
||||
|
||||
TIME_LENGTH = 16; % Dont Change this
|
||||
ScanTimes = zeros(nScans, 1);
|
||||
for i=1:nScans
|
||||
ithScanName = Scans{i};
|
||||
ithScanTime = str2double(ithScanName(1:TIME_LENGTH));
|
||||
ScanTimes(i) = ithScanTime;
|
||||
end
|
||||
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,9 +0,0 @@
|
|||
function [ Ptcloud ] = getNearestPtcloud( ScanTimes, curTime, Scans, ScanDir)
|
||||
|
||||
[MinTimeDelta, ArgminIdx] = min(abs(ScanTimes-curTime));
|
||||
ArgminBinName = Scans{ArgminIdx};
|
||||
ArgminBinPath = strcat(ScanDir, ArgminBinName);
|
||||
Ptcloud = NCLTbin2Ptcloud(ArgminBinPath);
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,23 +0,0 @@
|
|||
function [ PlaceIdx ] = getPlaceIdx(curPose, PlaceIndexAndGridCenters)
|
||||
|
||||
%% load meta file
|
||||
|
||||
%% Main
|
||||
curX = curPose(1);
|
||||
curY = curPose(2);
|
||||
|
||||
PlaceCellCenters = PlaceIndexAndGridCenters(:, 2:3);
|
||||
nPlaces = length(PlaceCellCenters);
|
||||
|
||||
Dists = zeros(nPlaces, 1);
|
||||
for ii=1:nPlaces
|
||||
Dist = norm(PlaceCellCenters(ii, :) - [curX, curY]);
|
||||
Dists(ii) = Dist;
|
||||
end
|
||||
|
||||
[NearestDist, NearestIdx] = min(Dists);
|
||||
|
||||
PlaceIdx = NearestIdx;
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,17 +0,0 @@
|
|||
function [ theta ] = getThetaFromXY( x, y )
|
||||
|
||||
if (x >= 0 && y >= 0)
|
||||
theta = 180/pi * atan(y/x);
|
||||
end
|
||||
if (x < 0 && y >= 0)
|
||||
theta = 180 - ((180/pi) * atan(y/(-x)));
|
||||
end
|
||||
if (x < 0 && y < 0)
|
||||
theta = 180 + ((180/pi) * atan(y/x));
|
||||
end
|
||||
if ( x >= 0 && y < 0)
|
||||
theta = 360 - ((180/pi) * atan((-y)/x));
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,28 +0,0 @@
|
|||
function [ PlaceIndexAndGridCenters ] = makeGridCellIndex ( xRange, yRange, PlaceCellSize )
|
||||
|
||||
xSize = xRange(2) - xRange(1);
|
||||
ySize = yRange(2) - yRange(1);
|
||||
|
||||
nGridX = round(xSize/PlaceCellSize);
|
||||
nGridY = round(ySize/PlaceCellSize);
|
||||
|
||||
xGridBoundaries = linspace(xRange(1), xRange(2), nGridX+1);
|
||||
yGridBoundaries = linspace(yRange(1), yRange(2), nGridY+1);
|
||||
|
||||
nTotalIndex = nGridX * nGridY;
|
||||
|
||||
curAssignedIndex = 1;
|
||||
PlaceIndexAndGridCenters = zeros(nTotalIndex, 3);
|
||||
for ii=1:nGridX
|
||||
xGridCenter = (xGridBoundaries(ii+1) + xGridBoundaries(ii))/2;
|
||||
for jj=1:nGridY
|
||||
yGridCenter = (yGridBoundaries(jj+1) + yGridBoundaries(jj))/2;
|
||||
|
||||
PlaceIndexAndGridCenters(curAssignedIndex, :) = [curAssignedIndex, xGridCenter, yGridCenter];
|
||||
curAssignedIndex = curAssignedIndex + 1;
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,22 +0,0 @@
|
|||
function [ ] = saveSCIcolor(curSCIcolor, DIR_SCIcolor, SamplingCounter, PlaceIdx, CellSize, ForB)
|
||||
|
||||
SCIcolor = curSCIcolor;
|
||||
|
||||
curSamplingCounter = num2str(SamplingCounter,'%0#6.f');
|
||||
curSamplingCounter = curSamplingCounter(1:end-1);
|
||||
|
||||
curPlaceIdx = num2str(PlaceIdx,'%0#6.f');
|
||||
curPlaceIdx = curPlaceIdx(1:end-1);
|
||||
|
||||
saveName = strcat(curSamplingCounter, '_', curPlaceIdx);
|
||||
saveDir = strcat(DIR_SCIcolor, CellSize, '/');
|
||||
if( ~exist(saveDir))
|
||||
mkdir(saveDir)
|
||||
end
|
||||
|
||||
savePath = strcat(saveDir, saveName, ForB, '.png');
|
||||
|
||||
imwrite(SCIcolor, savePath);
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,302 +0,0 @@
|
|||
{
|
||||
"cells": [
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 9,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"import os\n",
|
||||
"import numpy as np \n",
|
||||
"import pandas as pd \n",
|
||||
"import tensorflow as tf\n",
|
||||
"import keras\n",
|
||||
"import matplotlib.pyplot as plt"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 115,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Data info \n",
|
||||
"rootDir = '..your_data_path/'\n",
|
||||
"\n",
|
||||
"Dataset = 'NCLT'\n",
|
||||
"TrainOrTest = '/Train/'\n",
|
||||
"SequenceDate = '2012-01-15'\n",
|
||||
"\n",
|
||||
"SCImiddlePath = '/5. SCI_jet0to15_BackAug/'\n",
|
||||
"\n",
|
||||
"GridCellSize = '10'"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 116,
|
||||
"metadata": {},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"/media/gskim/IRAP-ADV1/Data/ICRA2019/NCLT/Train/2012-01-15/5. SCI_jet0to15_BackAug/10/\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"DataPath = rootDir + Dataset + TrainOrTest + SequenceDate + SCImiddlePath + GridCellSize + '/'\n",
|
||||
"print(DataPath)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 181,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"def getTrainingDataNCLT(DataPath, SequenceDate): \n",
|
||||
"\n",
|
||||
" # info\n",
|
||||
" WholeData = os.listdir(DataPath)\n",
|
||||
" nWholeData = len(WholeData)\n",
|
||||
" print(str(nWholeData) + ' data exist in ' + SequenceDate)\n",
|
||||
" \n",
|
||||
" # read \n",
|
||||
" X = []\n",
|
||||
" y = []\n",
|
||||
" for ii in range(nWholeData):\n",
|
||||
" dataName = WholeData[ii]\n",
|
||||
" dataPath = DataPath + dataName\n",
|
||||
" \n",
|
||||
" dataTrajNodeOrder = int(dataName[0:5])\n",
|
||||
"\n",
|
||||
" SCI = plt.imread(dataPath)\n",
|
||||
" dataPlaceIndex = int(dataName[6:11])\n",
|
||||
" \n",
|
||||
" X.append(SCI)\n",
|
||||
" y.append(dataPlaceIndex)\n",
|
||||
" \n",
|
||||
" # progress message \n",
|
||||
" if ii%1000==0:\n",
|
||||
" print(str(format((ii/nWholeData)*100, '.1f')), '% loaded.')\n",
|
||||
" \n",
|
||||
" \n",
|
||||
" dataShape = SCI.shape\n",
|
||||
" \n",
|
||||
" # X\n",
|
||||
" X_nd = np.zeros(shape=(nWholeData, dataShape[0], dataShape[1], dataShape[2]))\n",
|
||||
" for jj in range(len(X)):\n",
|
||||
" X_nd[jj, :, :] = X[jj]\n",
|
||||
" X_nd = X_nd.astype('float32')\n",
|
||||
" \n",
|
||||
" # y (one-hot encoded)\n",
|
||||
" from sklearn.preprocessing import LabelEncoder\n",
|
||||
" lbl_enc = LabelEncoder()\n",
|
||||
" lbl_enc.fit(y)\n",
|
||||
" \n",
|
||||
" ClassesTheSequenceHave = lbl_enc.classes_\n",
|
||||
" nClassesTheSequenceHave = len(ClassesTheSequenceHave)\n",
|
||||
" \n",
|
||||
" y = lbl_enc.transform(y)\n",
|
||||
" y_nd = keras.utils.np_utils.to_categorical(y, num_classes=nClassesTheSequenceHave)\n",
|
||||
"\n",
|
||||
" # log message \n",
|
||||
" print('Data size: %s' % nWholeData)\n",
|
||||
" print(' ')\n",
|
||||
" print('Data shape:', X_nd.shape)\n",
|
||||
" print('Label shape:', y_nd.shape)\n",
|
||||
" \n",
|
||||
" return X_nd, y_nd, lbl_enc\n",
|
||||
" "
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 182,
|
||||
"metadata": {
|
||||
"scrolled": false
|
||||
},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"15078 data exist in 2012-01-15\n",
|
||||
"0.0 % loaded.\n",
|
||||
"6.6 % loaded.\n",
|
||||
"13.3 % loaded.\n",
|
||||
"19.9 % loaded.\n",
|
||||
"26.5 % loaded.\n",
|
||||
"33.2 % loaded.\n",
|
||||
"39.8 % loaded.\n",
|
||||
"46.4 % loaded.\n",
|
||||
"53.1 % loaded.\n",
|
||||
"59.7 % loaded.\n",
|
||||
"66.3 % loaded.\n",
|
||||
"73.0 % loaded.\n",
|
||||
"79.6 % loaded.\n",
|
||||
"86.2 % loaded.\n",
|
||||
"92.9 % loaded.\n",
|
||||
"99.5 % loaded.\n",
|
||||
"Data size: 15078\n",
|
||||
"Data shape: (15078, 40, 120, 3)\n",
|
||||
"Label shape: (15078, 579)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"[X, y, lbl_enc] = getTrainingDataNCLT(DataPath, SequenceDate)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 172,
|
||||
"metadata": {},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"(40, 120, 3)\n",
|
||||
"(579,)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"dataShape = X[0].shape\n",
|
||||
"labelShape = y[0].shape\n",
|
||||
"\n",
|
||||
"print(dataShape)\n",
|
||||
"print(labelShape)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 173,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Model \n",
|
||||
"from keras import backend as K\n",
|
||||
"K.clear_session()\n",
|
||||
"\n",
|
||||
"ModelName = 'my_model'\n",
|
||||
"\n",
|
||||
"Drop1 = 0.7\n",
|
||||
"Drop2 = 0.7\n",
|
||||
"\n",
|
||||
"KernelSize = 5\n",
|
||||
"\n",
|
||||
"nConv1Filter = 64\n",
|
||||
"nConv2Filter = 128\n",
|
||||
"nConv3Filter = 256\n",
|
||||
"\n",
|
||||
"nFCN1 = 64\n",
|
||||
"\n",
|
||||
"inputs = keras.layers.Input(shape=(dataShape[0], dataShape[1], dataShape[2]))\n",
|
||||
"x = keras.layers.Conv2D(filters=nConv1Filter, kernel_size=KernelSize, activation='relu', padding='same')(inputs)\n",
|
||||
"x = keras.layers.MaxPooling2D(pool_size=(2, 2), strides=None, padding='valid')(x)\n",
|
||||
"x = keras.layers.BatchNormalization()(x)\n",
|
||||
"x = keras.layers.Conv2D(filters=nConv2Filter, kernel_size=KernelSize, activation='relu', padding='same')(x)\n",
|
||||
"x = keras.layers.MaxPool2D()(x)\n",
|
||||
"x = keras.layers.BatchNormalization()(x)\n",
|
||||
"x = keras.layers.Conv2D(filters=nConv3Filter, kernel_size=KernelSize, activation='relu', padding='same')(x)\n",
|
||||
"x = keras.layers.MaxPool2D()(x)\n",
|
||||
"x = keras.layers.Flatten()(x)\n",
|
||||
"x = keras.layers.Dropout(rate=Drop1)(x)\n",
|
||||
"x = keras.layers.Dense(units=nFCN1)(x)\n",
|
||||
"x = keras.layers.Dropout(rate=Drop2)(x)\n",
|
||||
"outputs = keras.layers.Dense(units=labelShape[0], activation='softmax')(x)\n",
|
||||
"\n",
|
||||
"model = keras.models.Model(inputs=inputs, outputs=outputs)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 174,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Model Compile \n",
|
||||
"model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['acc'])\n",
|
||||
"model.build(None,)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 1,
|
||||
"metadata": {
|
||||
"scrolled": true
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"#Train\n",
|
||||
"X_train = X\n",
|
||||
"y_train = y\n",
|
||||
"\n",
|
||||
"nEpoch = 200\n",
|
||||
"\n",
|
||||
"model.fit(X_train,\n",
|
||||
" y_train,\n",
|
||||
" epochs=nEpoch,\n",
|
||||
" batch_size=64,\n",
|
||||
" verbose=1\n",
|
||||
" )"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 2,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"#Train\n",
|
||||
"X_train = X\n",
|
||||
"y_train = y\n",
|
||||
"\n",
|
||||
"nEpoch = 200\n",
|
||||
"\n",
|
||||
"model.fit(X_train,\n",
|
||||
" y_train,\n",
|
||||
" epochs=nEpoch,\n",
|
||||
" batch_size=64,\n",
|
||||
" verbose=1\n",
|
||||
" )"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 186,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# model save \n",
|
||||
"modelName = 'ModelWS_' + GridCellSize + 'm_' + ModelName + '.h5'\n",
|
||||
"model.save(modelName)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"metadata": {
|
||||
"kernelspec": {
|
||||
"display_name": "Python 3",
|
||||
"language": "python",
|
||||
"name": "python3"
|
||||
},
|
||||
"language_info": {
|
||||
"codemirror_mode": {
|
||||
"name": "ipython",
|
||||
"version": 3
|
||||
},
|
||||
"file_extension": ".py",
|
||||
"mimetype": "text/x-python",
|
||||
"name": "python",
|
||||
"nbconvert_exporter": "python",
|
||||
"pygments_lexer": "ipython3",
|
||||
"version": "3.5.2"
|
||||
}
|
||||
},
|
||||
"nbformat": 4,
|
||||
"nbformat_minor": 2
|
||||
}
|
||||
|
|
@ -1,302 +0,0 @@
|
|||
{
|
||||
"cells": [
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 9,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"import os\n",
|
||||
"import numpy as np \n",
|
||||
"import pandas as pd \n",
|
||||
"import tensorflow as tf\n",
|
||||
"import keras\n",
|
||||
"import matplotlib.pyplot as plt"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 115,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Data info \n",
|
||||
"rootDir = '..your_data_path/'\n",
|
||||
"\n",
|
||||
"Dataset = 'NCLT'\n",
|
||||
"TrainOrTest = '/Train/'\n",
|
||||
"SequenceDate = '2012-01-15'\n",
|
||||
"\n",
|
||||
"SCImiddlePath = '/5. SCI_jet0to15_BackAug/'\n",
|
||||
"\n",
|
||||
"GridCellSize = '10'"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 116,
|
||||
"metadata": {},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"/media/gskim/IRAP-ADV1/Data/ICRA2019/NCLT/Train/2012-01-15/5. SCI_jet0to15_BackAug/10/\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"DataPath = rootDir + Dataset + TrainOrTest + SequenceDate + SCImiddlePath + GridCellSize + '/'\n",
|
||||
"print(DataPath)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 181,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"def getTrainingDataNCLT(DataPath, SequenceDate): \n",
|
||||
"\n",
|
||||
" # info\n",
|
||||
" WholeData = os.listdir(DataPath)\n",
|
||||
" nWholeData = len(WholeData)\n",
|
||||
" print(str(nWholeData) + ' data exist in ' + SequenceDate)\n",
|
||||
" \n",
|
||||
" # read \n",
|
||||
" X = []\n",
|
||||
" y = []\n",
|
||||
" for ii in range(nWholeData):\n",
|
||||
" dataName = WholeData[ii]\n",
|
||||
" dataPath = DataPath + dataName\n",
|
||||
" \n",
|
||||
" dataTrajNodeOrder = int(dataName[0:5])\n",
|
||||
"\n",
|
||||
" SCI = plt.imread(dataPath)\n",
|
||||
" dataPlaceIndex = int(dataName[6:11])\n",
|
||||
" \n",
|
||||
" X.append(SCI)\n",
|
||||
" y.append(dataPlaceIndex)\n",
|
||||
" \n",
|
||||
" # progress message \n",
|
||||
" if ii%1000==0:\n",
|
||||
" print(str(format((ii/nWholeData)*100, '.1f')), '% loaded.')\n",
|
||||
" \n",
|
||||
" \n",
|
||||
" dataShape = SCI.shape\n",
|
||||
" \n",
|
||||
" # X\n",
|
||||
" X_nd = np.zeros(shape=(nWholeData, dataShape[0], dataShape[1], dataShape[2]))\n",
|
||||
" for jj in range(len(X)):\n",
|
||||
" X_nd[jj, :, :] = X[jj]\n",
|
||||
" X_nd = X_nd.astype('float32')\n",
|
||||
" \n",
|
||||
" # y (one-hot encoded)\n",
|
||||
" from sklearn.preprocessing import LabelEncoder\n",
|
||||
" lbl_enc = LabelEncoder()\n",
|
||||
" lbl_enc.fit(y)\n",
|
||||
" \n",
|
||||
" ClassesTheSequenceHave = lbl_enc.classes_\n",
|
||||
" nClassesTheSequenceHave = len(ClassesTheSequenceHave)\n",
|
||||
" \n",
|
||||
" y = lbl_enc.transform(y)\n",
|
||||
" y_nd = keras.utils.np_utils.to_categorical(y, num_classes=nClassesTheSequenceHave)\n",
|
||||
"\n",
|
||||
" # log message \n",
|
||||
" print('Data size: %s' % nWholeData)\n",
|
||||
" print(' ')\n",
|
||||
" print('Data shape:', X_nd.shape)\n",
|
||||
" print('Label shape:', y_nd.shape)\n",
|
||||
" \n",
|
||||
" return X_nd, y_nd, lbl_enc\n",
|
||||
" "
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 182,
|
||||
"metadata": {
|
||||
"scrolled": false
|
||||
},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"15078 data exist in 2012-01-15\n",
|
||||
"0.0 % loaded.\n",
|
||||
"6.6 % loaded.\n",
|
||||
"13.3 % loaded.\n",
|
||||
"19.9 % loaded.\n",
|
||||
"26.5 % loaded.\n",
|
||||
"33.2 % loaded.\n",
|
||||
"39.8 % loaded.\n",
|
||||
"46.4 % loaded.\n",
|
||||
"53.1 % loaded.\n",
|
||||
"59.7 % loaded.\n",
|
||||
"66.3 % loaded.\n",
|
||||
"73.0 % loaded.\n",
|
||||
"79.6 % loaded.\n",
|
||||
"86.2 % loaded.\n",
|
||||
"92.9 % loaded.\n",
|
||||
"99.5 % loaded.\n",
|
||||
"Data size: 15078\n",
|
||||
"Data shape: (15078, 40, 120, 3)\n",
|
||||
"Label shape: (15078, 579)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"[X, y, lbl_enc] = getTrainingDataNCLT(DataPath, SequenceDate)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 172,
|
||||
"metadata": {},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"(40, 120, 3)\n",
|
||||
"(579,)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"dataShape = X[0].shape\n",
|
||||
"labelShape = y[0].shape\n",
|
||||
"\n",
|
||||
"print(dataShape)\n",
|
||||
"print(labelShape)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 173,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Model \n",
|
||||
"from keras import backend as K\n",
|
||||
"K.clear_session()\n",
|
||||
"\n",
|
||||
"ModelName = 'my_model'\n",
|
||||
"\n",
|
||||
"Drop1 = 0.7\n",
|
||||
"Drop2 = 0.7\n",
|
||||
"\n",
|
||||
"KernelSize = 5\n",
|
||||
"\n",
|
||||
"nConv1Filter = 64\n",
|
||||
"nConv2Filter = 128\n",
|
||||
"nConv3Filter = 256\n",
|
||||
"\n",
|
||||
"nFCN1 = 64\n",
|
||||
"\n",
|
||||
"inputs = keras.layers.Input(shape=(dataShape[0], dataShape[1], dataShape[2]))\n",
|
||||
"x = keras.layers.Conv2D(filters=nConv1Filter, kernel_size=KernelSize, activation='relu', padding='same')(inputs)\n",
|
||||
"x = keras.layers.MaxPooling2D(pool_size=(2, 2), strides=None, padding='valid')(x)\n",
|
||||
"x = keras.layers.BatchNormalization()(x)\n",
|
||||
"x = keras.layers.Conv2D(filters=nConv2Filter, kernel_size=KernelSize, activation='relu', padding='same')(x)\n",
|
||||
"x = keras.layers.MaxPool2D()(x)\n",
|
||||
"x = keras.layers.BatchNormalization()(x)\n",
|
||||
"x = keras.layers.Conv2D(filters=nConv3Filter, kernel_size=KernelSize, activation='relu', padding='same')(x)\n",
|
||||
"x = keras.layers.MaxPool2D()(x)\n",
|
||||
"x = keras.layers.Flatten()(x)\n",
|
||||
"x = keras.layers.Dropout(rate=Drop1)(x)\n",
|
||||
"x = keras.layers.Dense(units=nFCN1)(x)\n",
|
||||
"x = keras.layers.Dropout(rate=Drop2)(x)\n",
|
||||
"outputs = keras.layers.Dense(units=labelShape[0], activation='softmax')(x)\n",
|
||||
"\n",
|
||||
"model = keras.models.Model(inputs=inputs, outputs=outputs)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 174,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Model Compile \n",
|
||||
"model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['acc'])\n",
|
||||
"model.build(None,)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 1,
|
||||
"metadata": {
|
||||
"scrolled": true
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"#Train\n",
|
||||
"X_train = X\n",
|
||||
"y_train = y\n",
|
||||
"\n",
|
||||
"nEpoch = 200\n",
|
||||
"\n",
|
||||
"model.fit(X_train,\n",
|
||||
" y_train,\n",
|
||||
" epochs=nEpoch,\n",
|
||||
" batch_size=64,\n",
|
||||
" verbose=1\n",
|
||||
" )"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 2,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"#Train\n",
|
||||
"X_train = X\n",
|
||||
"y_train = y\n",
|
||||
"\n",
|
||||
"nEpoch = 200\n",
|
||||
"\n",
|
||||
"model.fit(X_train,\n",
|
||||
" y_train,\n",
|
||||
" epochs=nEpoch,\n",
|
||||
" batch_size=64,\n",
|
||||
" verbose=1\n",
|
||||
" )"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 186,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# model save \n",
|
||||
"modelName = 'ModelWS_' + GridCellSize + 'm_' + ModelName + '.h5'\n",
|
||||
"model.save(modelName)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"metadata": {
|
||||
"kernelspec": {
|
||||
"display_name": "Python 3",
|
||||
"language": "python",
|
||||
"name": "python3"
|
||||
},
|
||||
"language_info": {
|
||||
"codemirror_mode": {
|
||||
"name": "ipython",
|
||||
"version": 3
|
||||
},
|
||||
"file_extension": ".py",
|
||||
"mimetype": "text/x-python",
|
||||
"name": "python",
|
||||
"nbconvert_exporter": "python",
|
||||
"pygments_lexer": "ipython3",
|
||||
"version": "3.5.2"
|
||||
}
|
||||
},
|
||||
"nbformat": 4,
|
||||
"nbformat_minor": 2
|
||||
}
|
||||
|
|
@ -1,303 +0,0 @@
|
|||
{
|
||||
"cells": [
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 1,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"import os\n",
|
||||
"import numpy as np \n",
|
||||
"import pandas as pd \n",
|
||||
"import tensorflow as tf\n",
|
||||
"import keras\n",
|
||||
"import matplotlib.pyplot as plt"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 2,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Data info \n",
|
||||
"rootDir = '..your_path'\n",
|
||||
"\n",
|
||||
"Dataset = 'NCLT'\n",
|
||||
"TrainOrTest = '/Test/'\n",
|
||||
"SequenceDate = '2013-04-05'\n",
|
||||
"\n",
|
||||
"SCImiddlePath = '/4. SCI_jet0to15/'\n",
|
||||
"\n",
|
||||
"GridCellSize = '10'\n",
|
||||
"\n",
|
||||
"DataPath = rootDir + Dataset + TrainOrTest + SequenceDate + SCImiddlePath + GridCellSize + '/'\n",
|
||||
"print(DataPath)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 3,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"DataPath = ICRArootDir + Dataset + TrainOrTest + SequenceDate + SCImiddlePath + GridCellSize + '/'\n",
|
||||
"print(DataPath)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 4,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"def getTestDataNCLT(DataPath, SequenceDate, lbl_enc_train): \n",
|
||||
"\n",
|
||||
" # load training label encoding information \n",
|
||||
" from sklearn.preprocessing import LabelEncoder\n",
|
||||
" ClassesTrainingSequenceHave = lbl_enc_train.classes_\n",
|
||||
" nClassesTrainingSequenceHave = len(ClassesTrainingSequenceHave)\n",
|
||||
"\n",
|
||||
" # info\n",
|
||||
" WholeData = os.listdir(DataPath)\n",
|
||||
" nWholeData = len(WholeData)\n",
|
||||
" print(str(nWholeData) + ' data exist in ' + SequenceDate)\n",
|
||||
" \n",
|
||||
" # read \n",
|
||||
" X_seen = []\n",
|
||||
" y_seen = []\n",
|
||||
" X_unseen = []\n",
|
||||
" y_unseen = []\n",
|
||||
" \n",
|
||||
" for ii in range(nWholeData):\n",
|
||||
" dataName = WholeData[ii]\n",
|
||||
" dataPath = DataPath + dataName\n",
|
||||
" \n",
|
||||
" dataTrajNodeOrder = int(dataName[0:5])\n",
|
||||
"\n",
|
||||
" SCI = plt.imread(dataPath)\n",
|
||||
" dataPlaceIndex = int(dataName[6:11])\n",
|
||||
" \n",
|
||||
" # if label is in the train, then save into the seen (seen is only subset to be tested)\n",
|
||||
" if dataPlaceIndex in ClassesTrainingSequenceHave:\n",
|
||||
" X_seen.append(SCI)\n",
|
||||
" y_seen.append(dataPlaceIndex)\n",
|
||||
" else:\n",
|
||||
" X_unseen.append(SCI)\n",
|
||||
" y_unseen.append(dataPlaceIndex)\n",
|
||||
" \n",
|
||||
" # progress message \n",
|
||||
" if ii%1000==0:\n",
|
||||
" print(str(format((ii/nWholeData)*100, '.1f')), '% loaded.')\n",
|
||||
" \n",
|
||||
" dataShape = SCI.shape\n",
|
||||
" \n",
|
||||
" # X\n",
|
||||
" nSeenData = len(X_seen)\n",
|
||||
" X_nd = np.zeros(shape=(nSeenData, dataShape[0], dataShape[1], dataShape[2]))\n",
|
||||
" for jj in range(nSeenData):\n",
|
||||
" X_nd[jj, :, :] = X_seen[jj]\n",
|
||||
" X_nd = X_nd.astype('float32')\n",
|
||||
" \n",
|
||||
" # y (one-hot encoded) \n",
|
||||
" y_seen = lbl_enc_train.transform(y_seen)\n",
|
||||
" y_nd = keras.utils.np_utils.to_categorical(y_seen, num_classes=nClassesTrainingSequenceHave)\n",
|
||||
"\n",
|
||||
" # log message \n",
|
||||
" print('Data size: %s' % nWholeData)\n",
|
||||
" print('- Seen data: %s' % len(X_seen))\n",
|
||||
" print('- Uneen data: %s' % len(X_unseen))\n",
|
||||
" print(' ')\n",
|
||||
" print('Data shape:', X_nd.shape)\n",
|
||||
" print('Label shape:', y_nd.shape)\n",
|
||||
" \n",
|
||||
" return X_nd, y_nd, X_unseen, y_unseen\n",
|
||||
" "
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 63,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# load training label encoding information for discriminate seen/unseen of test \n",
|
||||
"import pickle\n",
|
||||
"TrainingDate = '2012-01-15'\n",
|
||||
"TrainingDataPath = 'data_pickle/Train_' + TrainingDate + '_SCI_color.pkl'\n",
|
||||
"\n",
|
||||
"with open(TrainingDataPath, 'rb') as f: # Python 3: open(..., 'rb')\n",
|
||||
" X_train, y_train, lbl_enc_train = pickle.load(f)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 4,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Load test data using training label encoder information \n",
|
||||
"[X_seen, y_seen, X_unseen, y_unseen] = getTestDataNCLT(DataPath, SequenceDate, lbl_enc_train)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 1,
|
||||
"metadata": {},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stderr",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"/home/gskim/anaconda3/envs/tfkeras/lib/python3.5/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n",
|
||||
" from ._conv import register_converters as _register_converters\n",
|
||||
"Using TensorFlow backend.\n"
|
||||
]
|
||||
},
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"_________________________________________________________________\n",
|
||||
"Layer (type) Output Shape Param # \n",
|
||||
"=================================================================\n",
|
||||
"input_1 (InputLayer) (None, 40, 120, 3) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"conv2d_1 (Conv2D) (None, 40, 120, 64) 4864 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"max_pooling2d_1 (MaxPooling2 (None, 20, 60, 64) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"batch_normalization_1 (Batch (None, 20, 60, 64) 256 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"conv2d_2 (Conv2D) (None, 20, 60, 128) 204928 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"max_pooling2d_2 (MaxPooling2 (None, 10, 30, 128) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"batch_normalization_2 (Batch (None, 10, 30, 128) 512 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"conv2d_3 (Conv2D) (None, 10, 30, 256) 819456 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"max_pooling2d_3 (MaxPooling2 (None, 5, 15, 256) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"flatten_1 (Flatten) (None, 19200) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dropout_1 (Dropout) (None, 19200) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dense_1 (Dense) (None, 64) 1228864 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dropout_2 (Dropout) (None, 64) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dense_2 (Dense) (None, 579) 37635 \n",
|
||||
"=================================================================\n",
|
||||
"Total params: 2,296,515\n",
|
||||
"Trainable params: 2,296,131\n",
|
||||
"Non-trainable params: 384\n",
|
||||
"_________________________________________________________________\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"from keras.models import load_model\n",
|
||||
"modelName = 'pre_trained_model/base0.h5'\n",
|
||||
"testModel = load_model(modelName)\n",
|
||||
"\n",
|
||||
"testModel.summary()"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 13,
|
||||
"metadata": {
|
||||
"scrolled": false
|
||||
},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"5170/5170 [==============================] - 1s 182us/step\n",
|
||||
"2012-02-04\n",
|
||||
"- Test score: 1.008079155962518\n",
|
||||
"- Test accuracy: 82.76595741913904\n",
|
||||
" \n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"# Load Trained net \n",
|
||||
"from keras.models import load_model\n",
|
||||
"modelName = 'model/base0.h5'\n",
|
||||
"testModel = load_model(modelName)\n",
|
||||
"\n",
|
||||
"# Predict \n",
|
||||
"scores_TEST = testModel.evaluate(X_seen, y_seen, verbose=1, batch_size=1000)\n",
|
||||
"print(SequenceDate)\n",
|
||||
"print('- Test score:', scores_TEST[0])\n",
|
||||
"print('- Test accuracy:', scores_TEST[1]*100)\n",
|
||||
"print(' ')"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 21,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# save prediction (for later top N analysis )\n",
|
||||
"y_seen_predicted = testModel.predict(X_seen)\n",
|
||||
"\n",
|
||||
"# save \n",
|
||||
"filename_y_seen_predicted = 'results_predictionvectors/base0/' + SequenceDate + '_seen_predicted'\n",
|
||||
"np.save(filename_y_seen_predicted, y_seen_predicted)\n",
|
||||
"\n",
|
||||
"# save prediction (for later top N analysis )\n",
|
||||
"X_unseen = np.array(X_unseen)\n",
|
||||
"y_unseen_predicted = testModel.predict(X_unseen)\n",
|
||||
"\n",
|
||||
"# save \n",
|
||||
"filename_y_unseen_predicted = 'results_predictionvectors/base0/' + SequenceDate + '_unseen_predicted'\n",
|
||||
"np.save(filename_y_unseen_predicted, y_unseen_predicted)\n"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 72,
|
||||
"metadata": {
|
||||
"scrolled": true
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"### save GT also \n",
|
||||
"\n",
|
||||
"# seen \n",
|
||||
"filename_y_seen_GT = 'results_predictionvectors/base0/' + SequenceDate + '_seen_GT'\n",
|
||||
"np.save(filename_y_seen_GT, y_seen)\n",
|
||||
"\n",
|
||||
"# unseen\n",
|
||||
"filename_y_unseen_GT = 'results_predictionvectors/base0/' + SequenceDate + '_unseen_GT'\n",
|
||||
"np.save(filename_y_unseen_GT, y_unseen)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"metadata": {
|
||||
"kernelspec": {
|
||||
"display_name": "Python 3",
|
||||
"language": "python",
|
||||
"name": "python3"
|
||||
},
|
||||
"language_info": {
|
||||
"codemirror_mode": {
|
||||
"name": "ipython",
|
||||
"version": 3
|
||||
},
|
||||
"file_extension": ".py",
|
||||
"mimetype": "text/x-python",
|
||||
"name": "python",
|
||||
"nbconvert_exporter": "python",
|
||||
"pygments_lexer": "ipython3",
|
||||
"version": "3.5.2"
|
||||
}
|
||||
},
|
||||
"nbformat": 4,
|
||||
"nbformat_minor": 2
|
||||
}
|
||||
|
|
@ -1,100 +0,0 @@
|
|||
clear
|
||||
|
||||
%% Path info
|
||||
Dataset = 'NCLT';
|
||||
Method = 'SCI';
|
||||
% ResultsDir = strcat('Result/', Dataset, '/', Method, '/');
|
||||
|
||||
ResultsDir = 'Result/';
|
||||
|
||||
%% Params
|
||||
FigIdx = 1;
|
||||
figure(FigIdx); clf;
|
||||
|
||||
TopNindexes = [25];
|
||||
nTopNindexes = length(TopNindexes);
|
||||
|
||||
%% Main
|
||||
SequenceNames = dir(ResultsDir); SequenceNames(1:2, :) = []; SequenceNames = {SequenceNames(:).name};
|
||||
nSequences = length(SequenceNames);
|
||||
|
||||
for ithTopN = 1:nTopNindexes
|
||||
|
||||
TopNidx = TopNindexes(ithTopN);
|
||||
|
||||
for ithSeq = 1:nSequences
|
||||
|
||||
% seq info
|
||||
ithSeqName = SequenceNames{ithSeq};
|
||||
ithSeqPath = strcat(ResultsDir, ithSeqName, '/');
|
||||
ithSeqPRcurveData = dir(ithSeqPath); ithSeqPRcurveData(1:2, :) = []; ithSeqPRcurveData = {ithSeqPRcurveData(:).name};
|
||||
|
||||
% load
|
||||
nCorrectRejectionsAll = load(strcat(ithSeqPath, ithSeqPRcurveData{1}));
|
||||
nCorrectRejectionsAll = nCorrectRejectionsAll.nCorrectRejections;
|
||||
nCorrectRejectionsForThisTopN = nCorrectRejectionsAll(TopNidx, :);
|
||||
|
||||
nFalseAlarmsAll = load(strcat(ithSeqPath, ithSeqPRcurveData{2}));
|
||||
nFalseAlarmsAll = nFalseAlarmsAll.nFalseAlarms;
|
||||
nFalseAlarmsForThisTopN = nFalseAlarmsAll(TopNidx, :);
|
||||
|
||||
nHitsAll = load(strcat(ithSeqPath, ithSeqPRcurveData{3}));
|
||||
nHitsAll = nHitsAll.nHits;
|
||||
nHitsForThisTopN = nHitsAll(TopNidx, :);
|
||||
|
||||
nMissesAll = load(strcat(ithSeqPath, ithSeqPRcurveData{4}));
|
||||
nMissesAll = nMissesAll.nMisses;
|
||||
nMissesForThisTopN = nMissesAll(TopNidx, :);
|
||||
|
||||
% info
|
||||
nTopNs = size(nCorrectRejectionsAll, 1);
|
||||
nThres = size(nCorrectRejectionsAll, 2);
|
||||
|
||||
% main
|
||||
Precisions = [];
|
||||
Recalls = [];
|
||||
Accuracies = [];
|
||||
for ithThres = 1:nThres
|
||||
nCorrectRejections = nCorrectRejectionsForThisTopN(ithThres);
|
||||
nFalseAlarms = nFalseAlarmsForThisTopN(ithThres);
|
||||
nHits = nHitsForThisTopN(ithThres);
|
||||
nMisses = nMissesForThisTopN(ithThres);
|
||||
|
||||
nTotalTestPlaces = nCorrectRejections + nFalseAlarms + nHits + nMisses;
|
||||
|
||||
Precision = nHits / (nHits + nFalseAlarms);
|
||||
Recall = nHits / (nHits + nMisses);
|
||||
Acc = (nHits + nCorrectRejections)/nTotalTestPlaces;
|
||||
|
||||
Precisions = [Precisions; Precision];
|
||||
Recalls = [Recalls; Recall];
|
||||
Accuracies = [Accuracies; Acc];
|
||||
end
|
||||
|
||||
% draw
|
||||
figure(FigIdx);
|
||||
plot(Recalls, Precisions, 'LineWidth', 2); % commonly x axis is recall
|
||||
title('SCI at NCLT');
|
||||
xlabel('Recall'); ylabel('Precision');
|
||||
% axis equal;
|
||||
xlim([0, 1]); ylim([0,1]);
|
||||
grid on; grid minor;
|
||||
hold on;
|
||||
|
||||
end
|
||||
|
||||
lgd = legend(SequenceNames, 'Location', 'best');
|
||||
lgd.FontSize = 9;
|
||||
lgd.FontWeight = 'bold';
|
||||
|
||||
|
||||
%% save
|
||||
% fileName = strcat('./results/', testDate, '_PRcurveWithEntropyThresVarying.png');
|
||||
% saveas(gcf, fileName)
|
||||
%
|
||||
% fileName = strcat('./results/', testDate, '_EntireWorkSpace.mat');
|
||||
% save(fileName)
|
||||
|
||||
end
|
||||
|
||||
|
||||
|
|
@ -1,142 +0,0 @@
|
|||
clear
|
||||
addpath(genpath('./'));
|
||||
|
||||
%% Setup
|
||||
Dataset = 'NCLT';
|
||||
Method = 'LearningSCI';
|
||||
ResultDir = strcat('/media/gskim/IRAP-ADV1/Data/ICRA2019/Experiments/#. trunk/', Dataset, '/', Method, '/10m/results_predictionvectors/base0/');
|
||||
|
||||
NCLTTestDateNames = {'2012-02-04', '2012-03-17', '2012-05-26', '2012-06-15', '2012-08-20', '2012-09-28', '2012-10-28', '2012-11-16', '2013-02-23', '2013-04-05'};
|
||||
nNCLTTestDates = length(NCLTTestDateNames);
|
||||
|
||||
|
||||
%% Main
|
||||
for ithDate = 1:nNCLTTestDates
|
||||
|
||||
TestDateName = NCLTTestDateNames{ithDate};
|
||||
|
||||
% path
|
||||
SeenGTs_Path = strcat(ResultDir, TestDateName, '_seen_GT.npy');
|
||||
SeenPredictions_Path = strcat(ResultDir, TestDateName, '_seen_predicted.npy');
|
||||
|
||||
UnseenGTs_Path = strcat(ResultDir, TestDateName, '_unseen_GT.npy');
|
||||
UnseenPredictions_Path = strcat(ResultDir, TestDateName, '_unseen_predicted.npy');
|
||||
|
||||
% load
|
||||
SeenGTs = double(readNPY(SeenGTs_Path));
|
||||
SeenPredictions = double(readNPY(SeenPredictions_Path));
|
||||
UnseenGTs = double(readNPY(UnseenGTs_Path));
|
||||
UnseenPredictions = double(readNPY(UnseenPredictions_Path));
|
||||
|
||||
% info
|
||||
nSeenPlaces = size(SeenGTs, 1);
|
||||
nUnseenPlaces = size(UnseenGTs, 1);
|
||||
|
||||
% concate seen and unseen for convenience
|
||||
% TotalGTs = [SeenGTs; UnseenGTs];
|
||||
TotalPredictions = [SeenPredictions; UnseenPredictions];
|
||||
TotalSeenFlags = [ ones(nSeenPlaces, 1); zeros(nUnseenPlaces, 1)]; % seen (1) or unseen (0)
|
||||
nTotalTestPlaces = nSeenPlaces + nUnseenPlaces;
|
||||
nTestData = nTotalTestPlaces;
|
||||
|
||||
|
||||
%% Main
|
||||
|
||||
% policy (top N)
|
||||
TopN = 25;
|
||||
TopNs = linspace(1, TopN, TopN);
|
||||
nTopNs = length(TopNs);
|
||||
|
||||
% Entropy thresholds
|
||||
Thresholds = linspace(0, 1, 200);
|
||||
nThresholds = length(Thresholds);
|
||||
|
||||
% Main variables to store the result for drawing PR curve
|
||||
nHits = zeros(nTopNs, nThresholds);
|
||||
nFalseAlarms = zeros(nTopNs, nThresholds);
|
||||
nCorrectRejections = zeros(nTopNs, nThresholds);
|
||||
nMisses = zeros(nTopNs, nThresholds);
|
||||
|
||||
for ith=1:nTestData
|
||||
tic
|
||||
|
||||
% Flag: Seen(1) or Unseen(0)
|
||||
SeenOrUnseen = TotalSeenFlags(ith);
|
||||
|
||||
% GT place idx (in this code, use it only for the seen case. thus knowing unseen place index is unnecessary in this case for PR curve analysis
|
||||
if(SeenOrUnseen == 1)
|
||||
[dummy, ithTruthPlaceIdx] = max(SeenGTs(ith, :)); % but this idx is not the real place index, plz refer the scikitlearn label maker
|
||||
else
|
||||
ithTruthPlaceIdx = nan;
|
||||
end
|
||||
|
||||
% ith prediction vector
|
||||
ithPrediction = TotalPredictions(ith, :);
|
||||
|
||||
% entropy
|
||||
EntropyOfPrediction = NormalizedEntropyOfVector(ithPrediction); % Entropy for module 1 (new place detection)
|
||||
|
||||
% top N predictions
|
||||
[NearestSoftmaxOutputs, idxs] = sort(ithPrediction, 'descend');
|
||||
NearestPlaceIdxs = idxs(1:TopN); % as mentioned above: this idx is not the real place index, plz refer the scikitlearn label maker
|
||||
|
||||
|
||||
for ithTopN = 1:nTopNs
|
||||
|
||||
ithNearestPlaceIdxs = NearestPlaceIdxs(1:ithTopN);
|
||||
|
||||
for ithThres = 1:nThresholds
|
||||
|
||||
ithDistThreshold = Thresholds(ithThres);
|
||||
|
||||
% main
|
||||
if(EntropyOfPrediction >= ithDistThreshold)
|
||||
% if over the theshold, it is considered unseen.
|
||||
% for unseen considered, no step 2 (recognition of place index), and just quit now.
|
||||
if(SeenOrUnseen == 0)
|
||||
% TN: Correct Rejection
|
||||
nCorrectRejections(ithTopN, ithThres) = nCorrectRejections(ithTopN, ithThres) + 1;
|
||||
else
|
||||
% FN: MISS
|
||||
nMisses(ithTopN, ithThres) = nMisses(ithTopN, ithThres) + 1;
|
||||
end
|
||||
else
|
||||
% if under the theshold, it is considered seen.
|
||||
% and then check the correctness
|
||||
if( ismember(ithTruthPlaceIdx, ithNearestPlaceIdxs) )
|
||||
% TP: Hit
|
||||
nHits(ithTopN, ithThres) = nHits(ithTopN, ithThres) + 1;
|
||||
else
|
||||
% FP: False Alarm
|
||||
nFalseAlarms(ithTopN, ithThres) = nFalseAlarms(ithTopN, ithThres) + 1;
|
||||
end
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
message = strcat(num2str(ith), "/", num2str(nTestData), " of ", TestDateName);
|
||||
disp(message);
|
||||
toc
|
||||
|
||||
end
|
||||
|
||||
|
||||
% save the results (PR curve is for the later.)
|
||||
savePath = strcat('Result/', TestDateName, '/');
|
||||
if(~(7==exist(savePath,'dir')))
|
||||
mkdir(savePath);
|
||||
end
|
||||
save(strcat(savePath, 'nCorrectRejections.mat'), 'nCorrectRejections');
|
||||
save(strcat(savePath, 'nMisses.mat'), 'nMisses');
|
||||
save(strcat(savePath, 'nHits.mat'), 'nHits');
|
||||
save(strcat(savePath, 'nFalseAlarms.mat'), 'nFalseAlarms');
|
||||
|
||||
end
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
Binary file not shown.
|
|
@ -1,16 +0,0 @@
|
|||
function H = EntropyOfVector(vec)
|
||||
|
||||
len = length(vec);
|
||||
|
||||
H = 0;
|
||||
for ith=1:len
|
||||
pith = vec(ith);
|
||||
if(pith == 0)
|
||||
H = H + 0; % 0log0 = 0
|
||||
else
|
||||
H = H + ( -1 * (pith*log2(pith)) );
|
||||
end
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,22 +0,0 @@
|
|||
function NormalizedH = NormalizedEntropyOfVector(vec)
|
||||
|
||||
len = length(vec);
|
||||
|
||||
% max entropy
|
||||
maxEntropy = -len*(1/len * log2(1/len));
|
||||
|
||||
H = 0;
|
||||
for ith=1:len
|
||||
pith = vec(ith);
|
||||
if(pith == 0)
|
||||
H = H + 0; % 0log0 = 0
|
||||
else
|
||||
H = H + ( -1 * (pith*log2(pith)) );
|
||||
end
|
||||
end
|
||||
|
||||
% return
|
||||
NormalizedH = H/maxEntropy;
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,303 +0,0 @@
|
|||
{
|
||||
"cells": [
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 1,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"import os\n",
|
||||
"import numpy as np \n",
|
||||
"import pandas as pd \n",
|
||||
"import tensorflow as tf\n",
|
||||
"import keras\n",
|
||||
"import matplotlib.pyplot as plt"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 2,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Data info \n",
|
||||
"rootDir = '..your_path'\n",
|
||||
"\n",
|
||||
"Dataset = 'NCLT'\n",
|
||||
"TrainOrTest = '/Test/'\n",
|
||||
"SequenceDate = '2013-04-05'\n",
|
||||
"\n",
|
||||
"SCImiddlePath = '/4. SCI_jet0to15/'\n",
|
||||
"\n",
|
||||
"GridCellSize = '10'\n",
|
||||
"\n",
|
||||
"DataPath = rootDir + Dataset + TrainOrTest + SequenceDate + SCImiddlePath + GridCellSize + '/'\n",
|
||||
"print(DataPath)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 3,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"DataPath = ICRArootDir + Dataset + TrainOrTest + SequenceDate + SCImiddlePath + GridCellSize + '/'\n",
|
||||
"print(DataPath)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 4,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"def getTestDataNCLT(DataPath, SequenceDate, lbl_enc_train): \n",
|
||||
"\n",
|
||||
" # load training label encoding information \n",
|
||||
" from sklearn.preprocessing import LabelEncoder\n",
|
||||
" ClassesTrainingSequenceHave = lbl_enc_train.classes_\n",
|
||||
" nClassesTrainingSequenceHave = len(ClassesTrainingSequenceHave)\n",
|
||||
"\n",
|
||||
" # info\n",
|
||||
" WholeData = os.listdir(DataPath)\n",
|
||||
" nWholeData = len(WholeData)\n",
|
||||
" print(str(nWholeData) + ' data exist in ' + SequenceDate)\n",
|
||||
" \n",
|
||||
" # read \n",
|
||||
" X_seen = []\n",
|
||||
" y_seen = []\n",
|
||||
" X_unseen = []\n",
|
||||
" y_unseen = []\n",
|
||||
" \n",
|
||||
" for ii in range(nWholeData):\n",
|
||||
" dataName = WholeData[ii]\n",
|
||||
" dataPath = DataPath + dataName\n",
|
||||
" \n",
|
||||
" dataTrajNodeOrder = int(dataName[0:5])\n",
|
||||
"\n",
|
||||
" SCI = plt.imread(dataPath)\n",
|
||||
" dataPlaceIndex = int(dataName[6:11])\n",
|
||||
" \n",
|
||||
" # if label is in the train, then save into the seen (seen is only subset to be tested)\n",
|
||||
" if dataPlaceIndex in ClassesTrainingSequenceHave:\n",
|
||||
" X_seen.append(SCI)\n",
|
||||
" y_seen.append(dataPlaceIndex)\n",
|
||||
" else:\n",
|
||||
" X_unseen.append(SCI)\n",
|
||||
" y_unseen.append(dataPlaceIndex)\n",
|
||||
" \n",
|
||||
" # progress message \n",
|
||||
" if ii%1000==0:\n",
|
||||
" print(str(format((ii/nWholeData)*100, '.1f')), '% loaded.')\n",
|
||||
" \n",
|
||||
" dataShape = SCI.shape\n",
|
||||
" \n",
|
||||
" # X\n",
|
||||
" nSeenData = len(X_seen)\n",
|
||||
" X_nd = np.zeros(shape=(nSeenData, dataShape[0], dataShape[1], dataShape[2]))\n",
|
||||
" for jj in range(nSeenData):\n",
|
||||
" X_nd[jj, :, :] = X_seen[jj]\n",
|
||||
" X_nd = X_nd.astype('float32')\n",
|
||||
" \n",
|
||||
" # y (one-hot encoded) \n",
|
||||
" y_seen = lbl_enc_train.transform(y_seen)\n",
|
||||
" y_nd = keras.utils.np_utils.to_categorical(y_seen, num_classes=nClassesTrainingSequenceHave)\n",
|
||||
"\n",
|
||||
" # log message \n",
|
||||
" print('Data size: %s' % nWholeData)\n",
|
||||
" print('- Seen data: %s' % len(X_seen))\n",
|
||||
" print('- Uneen data: %s' % len(X_unseen))\n",
|
||||
" print(' ')\n",
|
||||
" print('Data shape:', X_nd.shape)\n",
|
||||
" print('Label shape:', y_nd.shape)\n",
|
||||
" \n",
|
||||
" return X_nd, y_nd, X_unseen, y_unseen\n",
|
||||
" "
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 63,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# load training label encoding information for discriminate seen/unseen of test \n",
|
||||
"import pickle\n",
|
||||
"TrainingDate = '2012-01-15'\n",
|
||||
"TrainingDataPath = 'data_pickle/Train_' + TrainingDate + '_SCI_color.pkl'\n",
|
||||
"\n",
|
||||
"with open(TrainingDataPath, 'rb') as f: # Python 3: open(..., 'rb')\n",
|
||||
" X_train, y_train, lbl_enc_train = pickle.load(f)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 4,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# Load test data using training label encoder information \n",
|
||||
"[X_seen, y_seen, X_unseen, y_unseen] = getTestDataNCLT(DataPath, SequenceDate, lbl_enc_train)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 1,
|
||||
"metadata": {},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stderr",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"/home/gskim/anaconda3/envs/tfkeras/lib/python3.5/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n",
|
||||
" from ._conv import register_converters as _register_converters\n",
|
||||
"Using TensorFlow backend.\n"
|
||||
]
|
||||
},
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"_________________________________________________________________\n",
|
||||
"Layer (type) Output Shape Param # \n",
|
||||
"=================================================================\n",
|
||||
"input_1 (InputLayer) (None, 40, 120, 3) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"conv2d_1 (Conv2D) (None, 40, 120, 64) 4864 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"max_pooling2d_1 (MaxPooling2 (None, 20, 60, 64) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"batch_normalization_1 (Batch (None, 20, 60, 64) 256 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"conv2d_2 (Conv2D) (None, 20, 60, 128) 204928 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"max_pooling2d_2 (MaxPooling2 (None, 10, 30, 128) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"batch_normalization_2 (Batch (None, 10, 30, 128) 512 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"conv2d_3 (Conv2D) (None, 10, 30, 256) 819456 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"max_pooling2d_3 (MaxPooling2 (None, 5, 15, 256) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"flatten_1 (Flatten) (None, 19200) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dropout_1 (Dropout) (None, 19200) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dense_1 (Dense) (None, 64) 1228864 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dropout_2 (Dropout) (None, 64) 0 \n",
|
||||
"_________________________________________________________________\n",
|
||||
"dense_2 (Dense) (None, 579) 37635 \n",
|
||||
"=================================================================\n",
|
||||
"Total params: 2,296,515\n",
|
||||
"Trainable params: 2,296,131\n",
|
||||
"Non-trainable params: 384\n",
|
||||
"_________________________________________________________________\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"from keras.models import load_model\n",
|
||||
"modelName = 'pre_trained_model/base0.h5'\n",
|
||||
"testModel = load_model(modelName)\n",
|
||||
"\n",
|
||||
"testModel.summary()"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 13,
|
||||
"metadata": {
|
||||
"scrolled": false
|
||||
},
|
||||
"outputs": [
|
||||
{
|
||||
"name": "stdout",
|
||||
"output_type": "stream",
|
||||
"text": [
|
||||
"5170/5170 [==============================] - 1s 182us/step\n",
|
||||
"2012-02-04\n",
|
||||
"- Test score: 1.008079155962518\n",
|
||||
"- Test accuracy: 82.76595741913904\n",
|
||||
" \n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"source": [
|
||||
"# Load Trained net \n",
|
||||
"from keras.models import load_model\n",
|
||||
"modelName = 'model/base0.h5'\n",
|
||||
"testModel = load_model(modelName)\n",
|
||||
"\n",
|
||||
"# Predict \n",
|
||||
"scores_TEST = testModel.evaluate(X_seen, y_seen, verbose=1, batch_size=1000)\n",
|
||||
"print(SequenceDate)\n",
|
||||
"print('- Test score:', scores_TEST[0])\n",
|
||||
"print('- Test accuracy:', scores_TEST[1]*100)\n",
|
||||
"print(' ')"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 21,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# save prediction (for later top N analysis )\n",
|
||||
"y_seen_predicted = testModel.predict(X_seen)\n",
|
||||
"\n",
|
||||
"# save \n",
|
||||
"filename_y_seen_predicted = 'results_predictionvectors/base0/' + SequenceDate + '_seen_predicted'\n",
|
||||
"np.save(filename_y_seen_predicted, y_seen_predicted)\n",
|
||||
"\n",
|
||||
"# save prediction (for later top N analysis )\n",
|
||||
"X_unseen = np.array(X_unseen)\n",
|
||||
"y_unseen_predicted = testModel.predict(X_unseen)\n",
|
||||
"\n",
|
||||
"# save \n",
|
||||
"filename_y_unseen_predicted = 'results_predictionvectors/base0/' + SequenceDate + '_unseen_predicted'\n",
|
||||
"np.save(filename_y_unseen_predicted, y_unseen_predicted)\n"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": 72,
|
||||
"metadata": {
|
||||
"scrolled": true
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"### save GT also \n",
|
||||
"\n",
|
||||
"# seen \n",
|
||||
"filename_y_seen_GT = 'results_predictionvectors/base0/' + SequenceDate + '_seen_GT'\n",
|
||||
"np.save(filename_y_seen_GT, y_seen)\n",
|
||||
"\n",
|
||||
"# unseen\n",
|
||||
"filename_y_unseen_GT = 'results_predictionvectors/base0/' + SequenceDate + '_unseen_GT'\n",
|
||||
"np.save(filename_y_unseen_GT, y_unseen)\n"
|
||||
]
|
||||
}
|
||||
],
|
||||
"metadata": {
|
||||
"kernelspec": {
|
||||
"display_name": "Python 3",
|
||||
"language": "python",
|
||||
"name": "python3"
|
||||
},
|
||||
"language_info": {
|
||||
"codemirror_mode": {
|
||||
"name": "ipython",
|
||||
"version": 3
|
||||
},
|
||||
"file_extension": ".py",
|
||||
"mimetype": "text/x-python",
|
||||
"name": "python",
|
||||
"nbconvert_exporter": "python",
|
||||
"pygments_lexer": "ipython3",
|
||||
"version": "3.5.2"
|
||||
}
|
||||
},
|
||||
"nbformat": 4,
|
||||
"nbformat_minor": 2
|
||||
}
|
||||
Binary file not shown.
Binary file not shown.
|
Before Width: | Height: | Size: 103 KiB |
|
|
@ -1,3 +0,0 @@
|
|||
function dist = DistBtn2Dpose(pose1, pose2)
|
||||
dist = sqrt( (pose1(1) - pose2(1))^2 + (pose1(2) - pose2(2))^2);
|
||||
end
|
||||
Binary file not shown.
|
|
@ -1,21 +0,0 @@
|
|||
function ptcloud = KITTIbin2PtcloudWithIndex(base_dir, index)
|
||||
|
||||
%% File path
|
||||
if( length(num2str(index)) == 4 )
|
||||
bin_path = strcat(base_dir, '00', num2str(index), '.bin');
|
||||
elseif (length(num2str(index)) == 3)
|
||||
bin_path = strcat(base_dir, '000', num2str(index), '.bin');
|
||||
elseif (length(num2str(index)) == 2)
|
||||
bin_path = strcat(base_dir, '0000', num2str(index), '.bin');
|
||||
elseif (length(num2str(index)) == 1)
|
||||
bin_path = strcat(base_dir, '00000', num2str(index), '.bin');
|
||||
end
|
||||
|
||||
%% Read
|
||||
fid = fopen(bin_path, 'rb'); raw_data = fread(fid, [4 inf], 'single'); fclose(fid);
|
||||
points = raw_data(1:3,:)';
|
||||
points(:, 3) = points(:, 3) + 1.9; % z in car coord.
|
||||
|
||||
ptcloud = pointCloud(points);
|
||||
|
||||
end % end of function
|
||||
|
|
@ -1,105 +0,0 @@
|
|||
clear; clc;
|
||||
addpath(genpath('../../../../matlab/'));
|
||||
|
||||
%% Change this to your path
|
||||
% vel_dir = '/media/gskim/Data/KITTI odo/data_odometry_velodyne/dataset/sequences/00/velodyne/';
|
||||
vel_dir = 'your_pointcloud_files_path';
|
||||
|
||||
|
||||
%% Params
|
||||
% below 3 parameters: same setting as the original paper (G. Kim, 18 IROS)
|
||||
max_range = 80; % meter
|
||||
num_sectors = 60;
|
||||
num_rings = 20;
|
||||
|
||||
num_candidates = 50; % means Scan Context-50 in the paper.
|
||||
|
||||
loop_thres = 0.2; % ### this is a user parameter ###
|
||||
|
||||
num_enough_node_diff = 50;
|
||||
|
||||
|
||||
%% Main
|
||||
load('GTposes.mat');
|
||||
|
||||
ringkeys = [];
|
||||
scancontexts = {};
|
||||
|
||||
loop_log = [];
|
||||
num_nodes = length(GTposes);
|
||||
for ith_node = 1:num_nodes-1
|
||||
|
||||
% information
|
||||
idx_query = ith_node;
|
||||
ptcloud = KITTIbin2PtcloudWithIndex(vel_dir, idx_query);
|
||||
|
||||
sc_query = Ptcloud2ScanContext(ptcloud, num_sectors, num_rings, max_range);
|
||||
scancontexts{end+1} = sc_query; % save into database
|
||||
|
||||
% ringkey tree
|
||||
ringkey = ScanContext2RingKey(sc_query);
|
||||
ringkeys = [ringkeys; ringkey];
|
||||
tree = createns(ringkeys, 'NSMethod', 'kdtree'); % Create object to use in k-nearest neighbor search
|
||||
|
||||
% try loop-detection after enough moving
|
||||
if(ith_node < num_candidates)
|
||||
continue;
|
||||
end
|
||||
|
||||
% find nearest candidates
|
||||
candidates = knnsearch(tree, ringkey, 'K', num_candidates);
|
||||
|
||||
% check dist btn candidates is lower than the given threshold.
|
||||
idx_nearest = 0;
|
||||
min_dist = inf; % initialization
|
||||
for ith_candidate = 1:length(candidates)
|
||||
|
||||
idx_candidate = candidates(ith_candidate);
|
||||
sc_candidate = scancontexts{idx_candidate};
|
||||
|
||||
% skip condition: do not check nearest measurements
|
||||
if( abs(idx_query - idx_candidate) < num_enough_node_diff)
|
||||
continue;
|
||||
end
|
||||
|
||||
% Main
|
||||
distance_to_query = DistanceBtnScanContexts(sc_query, sc_candidate);
|
||||
if( distance_to_query > loop_thres)
|
||||
continue;
|
||||
end
|
||||
|
||||
if( distance_to_query < min_dist)
|
||||
idx_nearest = idx_candidate;
|
||||
min_dist = distance_to_query;
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
% log the result
|
||||
if( ~isequal(min_dist, inf)) % that is, when any loop (satisfied under acceptance theshold) occured.
|
||||
pose_dist_real = DistBtn2Dpose(GTposes(idx_query,:), GTposes(idx_nearest,:));
|
||||
loop_log = [loop_log; ...
|
||||
idx_query, idx_nearest, min_dist, pose_dist_real];
|
||||
end
|
||||
|
||||
%% Log Message: procedure and LoopFound Event
|
||||
if( rem(idx_query, 100) == 0)
|
||||
disp( strcat(num2str(idx_query), '.bin processed') );
|
||||
end
|
||||
if( ~isequal(min_dist, inf) )
|
||||
disp( strcat("Loop found: ", num2str(idx_query), " <-> ", num2str(idx_nearest), " with dist ", num2str(min_dist)));
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
||||
%% Save the result into csv file
|
||||
save_dir = strcat('./result/', num2str(num_candidates),'/', num2str(loop_thres), '/');
|
||||
save_file_name = strcat(save_dir, '/LogLoopFound.csv');
|
||||
if( ~exist(save_dir))
|
||||
mkdir(save_dir)
|
||||
end
|
||||
csvwrite(save_file_name, loop_log);
|
||||
|
||||
|
||||
|
||||
|
|
@ -1,757 +0,0 @@
|
|||
1568,119,0.18125,2.3459
|
||||
1569,121,0.15028,2.0697
|
||||
1570,123,0.18087,1.8398
|
||||
1571,123,0.17516,1.7656
|
||||
1572,126,0.1617,1.6084
|
||||
1573,126,0.12506,1.5125
|
||||
1574,126,0.13887,1.6024
|
||||
1575,129,0.14479,1.2933
|
||||
1576,130,0.11332,1.2109
|
||||
1577,131,0.10118,1.1562
|
||||
1578,132,0.11642,1.1173
|
||||
1579,133,0.14223,1.0887
|
||||
1580,134,0.13837,1.0587
|
||||
1581,136,0.15035,0.96174
|
||||
1582,136,0.13031,1.0202
|
||||
1583,138,0.14318,0.84406
|
||||
1584,140,0.1356,1.0254
|
||||
1585,140,0.1024,0.75281
|
||||
1586,141,0.11191,0.71942
|
||||
1587,142,0.11754,0.68515
|
||||
1588,143,0.1062,0.6584
|
||||
1589,144,0.098269,0.62726
|
||||
1590,145,0.093211,0.60696
|
||||
1591,146,0.088597,0.59144
|
||||
1592,147,0.11178,0.58298
|
||||
1593,148,0.088569,0.58158
|
||||
1594,149,0.090449,0.5925
|
||||
1595,150,0.075444,0.6126
|
||||
1596,151,0.071824,0.64588
|
||||
1597,152,0.091617,0.68911
|
||||
1598,153,0.082827,0.73222
|
||||
1599,154,0.11145,0.78655
|
||||
1600,156,0.10101,0.51769
|
||||
1601,156,0.085753,0.89851
|
||||
1602,157,0.074695,0.95616
|
||||
1603,159,0.060621,0.57845
|
||||
1604,160,0.036375,0.59372
|
||||
1605,161,0.030381,0.60652
|
||||
1606,162,0.048665,0.61569
|
||||
1607,163,0.062883,0.61811
|
||||
1608,164,0.043436,0.61427
|
||||
1609,165,0.090096,0.60787
|
||||
1610,166,0.11956,0.60152
|
||||
1611,167,0.10092,0.59354
|
||||
1612,168,0.12087,0.59199
|
||||
1613,170,0.098452,0.61371
|
||||
1614,170,0.11376,0.59625
|
||||
1615,172,0.10143,0.51073
|
||||
1616,172,0.10065,0.63149
|
||||
1617,174,0.10071,0.40156
|
||||
1618,175,0.066769,0.35817
|
||||
1619,176,0.088933,0.33
|
||||
1620,177,0.11702,0.34074
|
||||
1621,178,0.10745,0.39436
|
||||
1622,180,0.13317,0.57002
|
||||
1623,181,0.13514,0.49705
|
||||
1624,182,0.13071,0.47461
|
||||
1625,183,0.12965,0.52818
|
||||
1626,185,0.12495,0.67878
|
||||
1627,186,0.10545,0.63886
|
||||
1628,187,0.12124,0.6919
|
||||
1629,189,0.092797,0.73359
|
||||
1630,191,0.093877,0.8266
|
||||
1631,193,0.11648,0.88208
|
||||
1632,194,0.088575,0.62795
|
||||
1633,196,0.059866,0.54343
|
||||
1634,198,0.056973,0.49308
|
||||
1635,199,0.11314,0.12862
|
||||
1636,201,0.14971,0.59225
|
||||
1637,200,0.19476,1.3008
|
||||
2442,390,0.16356,1.4154
|
||||
2443,390,0.14665,1.0374
|
||||
2444,391,0.14361,0.82121
|
||||
2445,392,0.16731,0.65271
|
||||
2446,393,0.16125,0.51617
|
||||
2447,394,0.12566,0.40868
|
||||
2448,395,0.17469,0.34193
|
||||
2449,396,0.092845,0.29563
|
||||
2450,397,0.10571,0.25573
|
||||
2451,397,0.17995,0.44304
|
||||
2452,398,0.19878,0.50376
|
||||
2453,400,0.19093,0.12883
|
||||
2454,401,0.1413,0.13343
|
||||
2455,403,0.088901,0.46873
|
||||
2456,404,0.095194,0.32575
|
||||
2457,405,0.16198,0.18097
|
||||
2458,408,0.19245,0.9109
|
||||
2459,409,0.11232,0.65213
|
||||
2460,410,0.15126,0.38529
|
||||
2461,412,0.094598,0.62195
|
||||
2462,412,0.1364,0.30654
|
||||
2463,414,0.17397,0.50585
|
||||
3285,2347,0.19181,1.7119
|
||||
3286,2348,0.18886,1.4355
|
||||
3287,2348,0.19508,1.3702
|
||||
3288,2350,0.15239,1.0513
|
||||
3289,2350,0.15637,0.82514
|
||||
3290,2351,0.10586,0.66724
|
||||
3291,2351,0.13678,0.69258
|
||||
3292,2352,0.13644,0.45817
|
||||
3293,2353,0.087613,0.31482
|
||||
3294,2353,0.15271,0.57803
|
||||
3295,2354,0.10603,0.35358
|
||||
3296,2355,0.045707,0.15249
|
||||
3297,2356,0.083127,0.1477
|
||||
3298,2356,0.12741,0.53191
|
||||
3299,2357,0.15497,0.3257
|
||||
3300,2358,0.14542,0.11591
|
||||
3301,2359,0.13331,0.082783
|
||||
3302,2359,0.10406,0.63607
|
||||
3303,2360,0.092642,0.46669
|
||||
3304,2361,0.094625,0.30149
|
||||
3305,2362,0.081233,0.14712
|
||||
3306,2363,0.071525,0.01429
|
||||
3307,2364,0.079,0.14063
|
||||
3308,2365,0.10557,0.28989
|
||||
3309,2365,0.086534,0.559
|
||||
3310,2366,0.078621,0.44814
|
||||
3311,2367,0.045815,0.35524
|
||||
3312,2368,0.055051,0.27569
|
||||
3313,2369,0.059123,0.22411
|
||||
3314,2370,0.066648,0.20413
|
||||
3315,2371,0.079022,0.21808
|
||||
3316,2372,0.081073,0.2491
|
||||
3317,2373,0.084914,0.29572
|
||||
3318,2374,0.1,0.34206
|
||||
3319,2375,0.090124,0.39653
|
||||
3320,2376,0.098208,0.44958
|
||||
3321,2377,0.085425,0.49533
|
||||
3322,2378,0.074547,0.53816
|
||||
3323,2379,0.08426,0.58167
|
||||
3324,2380,0.08894,0.62385
|
||||
3325,2381,0.078235,0.66161
|
||||
3326,2382,0.091711,0.69853
|
||||
3327,2383,0.058422,0.72976
|
||||
3328,2384,0.066054,0.74093
|
||||
3329,2385,0.064545,0.74904
|
||||
3330,2386,0.067067,0.73265
|
||||
3331,2387,0.061414,0.71872
|
||||
3332,2388,0.090268,0.70308
|
||||
3333,2389,0.078915,0.69906
|
||||
3334,2390,0.083742,0.69499
|
||||
3335,2391,0.058813,0.67748
|
||||
3336,2392,0.089391,0.66885
|
||||
3337,2393,0.052395,0.66004
|
||||
3338,2394,0.066151,0.65691
|
||||
3339,2395,0.080086,0.65148
|
||||
3340,2396,0.059779,0.65392
|
||||
3341,2397,0.062375,0.65045
|
||||
3342,2398,0.073409,0.64607
|
||||
3343,2399,0.071161,0.64021
|
||||
3344,2400,0.050122,0.64122
|
||||
3345,2401,0.038577,0.6421
|
||||
3346,2402,0.054643,0.65593
|
||||
3347,2403,0.068992,0.66919
|
||||
3348,2404,0.062084,0.70059
|
||||
3349,2405,0.068393,0.72263
|
||||
3350,2406,0.094639,0.75104
|
||||
3351,2407,0.083902,0.77198
|
||||
3352,2408,0.08078,0.79738
|
||||
3353,2409,0.085166,0.81228
|
||||
3354,2410,0.079796,0.83729
|
||||
3355,2411,0.070817,0.85167
|
||||
3356,2412,0.099746,0.84575
|
||||
3357,2413,0.078259,0.83719
|
||||
3358,2414,0.087562,0.83061
|
||||
3359,2415,0.09848,0.8226
|
||||
3360,2416,0.080679,0.81034
|
||||
3361,2416,0.087786,1.0043
|
||||
3362,2417,0.085017,0.93943
|
||||
3363,2418,0.077789,0.85353
|
||||
3364,2419,0.098126,0.78161
|
||||
3365,2420,0.11023,0.70018
|
||||
3366,2420,0.11876,0.9617
|
||||
3367,2421,0.089966,0.84369
|
||||
3368,2422,0.1494,0.73867
|
||||
3369,2422,0.11137,1.1043
|
||||
3370,2423,0.11026,0.99525
|
||||
3371,2424,0.089247,0.89416
|
||||
3372,2425,0.10631,0.78985
|
||||
3373,2425,0.11679,1.1914
|
||||
3374,2426,0.091945,1.0832
|
||||
3375,2427,0.086268,0.97989
|
||||
3376,2428,0.049368,0.88783
|
||||
3377,2429,0.072314,0.80638
|
||||
3378,2430,0.052582,0.69987
|
||||
3379,2431,0.094895,0.59817
|
||||
3380,2431,0.10076,1.0012
|
||||
3381,2432,0.1007,0.9295
|
||||
3382,2433,0.049914,0.85069
|
||||
3383,2434,0.045934,0.74765
|
||||
3384,2435,0.069366,0.69214
|
||||
3385,2436,0.066831,0.61126
|
||||
3386,2437,0.096679,0.53902
|
||||
3387,2438,0.097736,0.45893
|
||||
3388,2438,0.099658,0.96359
|
||||
3389,2439,0.088638,0.91281
|
||||
3390,2440,0.081316,0.88109
|
||||
3391,2441,0.079838,0.78725
|
||||
3392,2442,0.12346,0.70026
|
||||
3393,2443,0.087868,0.62774
|
||||
3394,2443,0.10694,1.1528
|
||||
3395,2444,0.0867,1.0204
|
||||
3396,2445,0.08955,0.87821
|
||||
3397,2446,0.086738,0.74263
|
||||
3398,2447,0.097434,0.58631
|
||||
3399,2447,0.067854,1.094
|
||||
3400,2448,0.054073,0.94282
|
||||
3401,2449,0.082468,0.78407
|
||||
3402,396,0.078349,0.9965
|
||||
3403,2450,0.10656,1.1245
|
||||
3404,2451,0.10494,0.96157
|
||||
3405,2452,0.089806,0.78443
|
||||
3406,2453,0.11585,0.60537
|
||||
3407,2453,0.082687,1.1265
|
||||
3408,2454,0.070972,0.94199
|
||||
3409,2455,0.083759,0.75355
|
||||
3410,2455,0.070209,1.285
|
||||
3411,2456,0.10177,1.098
|
||||
3412,405,0.12,0.89414
|
||||
3413,406,0.07472,0.91874
|
||||
3414,407,0.12934,0.94777
|
||||
3415,2459,0.095186,1.0739
|
||||
3416,409,0.14754,1.0207
|
||||
3417,411,0.12847,0.60378
|
||||
3418,2461,0.11527,1.2465
|
||||
3419,413,0.10909,0.69339
|
||||
3420,414,0.095259,0.71018
|
||||
3421,415,0.084456,0.70203
|
||||
3422,416,0.10397,0.7228
|
||||
3423,417,0.07364,0.73688
|
||||
3424,418,0.10099,0.76916
|
||||
3425,419,0.094113,0.79142
|
||||
3426,422,0.11695,0.11838
|
||||
3427,421,0.11265,0.86118
|
||||
3428,424,0.12608,0.2286
|
||||
3429,424,0.098531,0.52433
|
||||
3430,425,0.074338,0.58793
|
||||
3431,426,0.066058,0.66997
|
||||
3432,427,0.07593,0.68653
|
||||
3433,430,0.076777,0.43547
|
||||
3434,431,0.10626,0.50459
|
||||
3435,431,0.090603,0.57493
|
||||
3436,432,0.084591,0.62021
|
||||
3437,433,0.06518,0.65516
|
||||
3438,434,0.088447,0.67053
|
||||
3439,435,0.084382,0.69085
|
||||
3440,437,0.08864,0.5711
|
||||
3441,438,0.071889,0.58659
|
||||
3442,439,0.06133,0.59776
|
||||
3443,439,0.076491,0.7442
|
||||
3444,441,0.083076,0.59403
|
||||
3445,442,0.053194,0.60042
|
||||
3446,443,0.043822,0.58572
|
||||
3447,444,0.06208,0.57422
|
||||
3448,445,0.052252,0.56499
|
||||
3449,446,0.054012,0.5492
|
||||
3450,447,0.047266,0.54206
|
||||
3451,448,0.043386,0.537
|
||||
3452,449,0.046498,0.53628
|
||||
3453,450,0.051607,0.55481
|
||||
3454,451,0.04954,0.61125
|
||||
3455,452,0.055524,0.67392
|
||||
3456,453,0.049512,0.72452
|
||||
3457,454,0.060479,0.78121
|
||||
3458,455,0.048154,0.82682
|
||||
3459,456,0.037072,0.86692
|
||||
3460,457,0.04674,0.90879
|
||||
3461,458,0.031691,0.94834
|
||||
3462,459,0.039648,0.99626
|
||||
3463,460,0.047547,1.0273
|
||||
3464,461,0.057568,1.0677
|
||||
3465,463,0.061845,0.90718
|
||||
3466,463,0.064674,1.1651
|
||||
3467,465,0.072989,0.88811
|
||||
3468,466,0.075313,0.89524
|
||||
3469,467,0.044292,0.92334
|
||||
3470,468,0.032245,0.98216
|
||||
3471,469,0.053995,1.0397
|
||||
3472,471,0.040517,0.85185
|
||||
3473,472,0.059152,0.82555
|
||||
3474,473,0.039805,0.85778
|
||||
3475,474,0.037291,0.92548
|
||||
3476,475,0.063179,1.0448
|
||||
3477,477,0.05835,0.79219
|
||||
3478,478,0.017437,0.81236
|
||||
3479,479,0.064769,0.8788
|
||||
3480,481,0.067249,0.75607
|
||||
3481,482,0.049923,0.71111
|
||||
3482,483,0.04944,0.73929
|
||||
3483,484,0.084984,0.83942
|
||||
3484,486,0.068865,0.66955
|
||||
3485,487,0.080969,0.645
|
||||
3486,488,0.096104,0.70935
|
||||
3487,490,0.079273,0.62396
|
||||
3488,491,0.070285,0.55952
|
||||
3489,492,0.081309,0.59844
|
||||
3490,494,0.079061,0.59827
|
||||
3491,495,0.038021,0.50946
|
||||
3492,496,0.041706,0.52806
|
||||
3493,498,0.074656,0.64311
|
||||
3494,499,0.069889,0.54968
|
||||
3495,500,0.073375,0.54568
|
||||
3496,501,0.066413,0.62479
|
||||
3497,502,0.066331,0.75863
|
||||
3498,504,0.04612,0.5742
|
||||
3499,505,0.083456,0.61658
|
||||
3500,507,0.072278,0.64292
|
||||
3501,508,0.06483,0.59317
|
||||
3502,509,0.045953,0.62332
|
||||
3503,511,0.071982,0.63206
|
||||
3504,512,0.051929,0.57741
|
||||
3505,513,0.062702,0.59741
|
||||
3506,514,0.097461,0.67465
|
||||
3507,516,0.08499,0.54534
|
||||
3508,517,0.08652,0.56008
|
||||
3509,519,0.10472,0.50263
|
||||
3510,520,0.1377,0.46874
|
||||
3511,521,0.11436,0.50328
|
||||
3512,522,0.13331,0.57529
|
||||
3513,524,0.14193,0.40398
|
||||
3514,524,0.12945,0.75625
|
||||
3515,526,0.16611,0.51414
|
||||
3516,528,0.13532,0.33883
|
||||
3517,529,0.11522,0.38343
|
||||
3518,530,0.097582,0.44403
|
||||
3519,530,0.10195,0.77263
|
||||
3520,533,0.10225,0.39314
|
||||
3521,533,0.087222,0.67149
|
||||
3522,551,0.093044,0.22042
|
||||
3523,551,0.077817,0.26192
|
||||
3524,561,0.10284,0.25655
|
||||
3525,563,0.092164,0.30675
|
||||
3526,566,0.089572,0.18999
|
||||
3527,567,0.095544,0.21718
|
||||
3528,569,0.099764,0.17982
|
||||
3529,570,0.10202,0.1727
|
||||
3530,571,0.12059,0.17599
|
||||
3531,572,0.088523,0.18541
|
||||
3532,573,0.083859,0.19611
|
||||
3533,573,0.092266,0.22882
|
||||
3534,574,0.065916,0.2085
|
||||
3535,572,0.075181,0.86744
|
||||
3536,573,0.086694,0.86023
|
||||
3537,577,0.09177,0.16811
|
||||
3538,575,0.07192,0.88213
|
||||
3539,579,0.098566,0.20077
|
||||
3540,577,0.11658,0.94366
|
||||
3541,579,0.10608,0.68274
|
||||
3542,581,0.10241,0.39468
|
||||
3543,582,0.079983,0.42449
|
||||
3544,583,0.09799,0.46025
|
||||
3545,584,0.065836,0.46168
|
||||
3546,585,0.060715,0.53953
|
||||
3547,586,0.078283,0.58405
|
||||
3548,587,0.074035,0.62879
|
||||
3549,588,0.092114,0.67884
|
||||
3550,589,0.099951,0.73316
|
||||
3551,590,0.092961,0.78744
|
||||
3552,592,0.11666,0.37682
|
||||
3553,592,0.11467,0.87151
|
||||
3554,594,0.089423,0.48545
|
||||
3555,595,0.048698,0.56237
|
||||
3556,596,0.030646,0.5912
|
||||
3557,597,0.092709,0.65082
|
||||
3558,599,0.076415,0.3088
|
||||
3559,600,0.064109,0.32145
|
||||
3560,601,0.092458,0.33833
|
||||
3561,602,0.087178,0.35998
|
||||
3562,603,0.065431,0.39903
|
||||
3563,604,0.079738,0.45687
|
||||
3564,605,0.068067,0.52619
|
||||
3565,606,0.065775,0.60701
|
||||
3566,608,0.069198,0.29107
|
||||
3567,609,0.042854,0.26908
|
||||
3568,609,0.073597,0.90412
|
||||
3569,610,0.07436,1.0097
|
||||
3570,612,0.027189,0.38187
|
||||
3571,613,0.03542,0.46832
|
||||
3572,614,0.054128,0.57312
|
||||
3573,615,0.058842,0.69129
|
||||
3574,616,0.088969,0.82029
|
||||
3575,618,0.068361,0.27177
|
||||
3576,619,0.07929,0.3252
|
||||
3577,620,0.061089,0.4271
|
||||
3578,621,0.087537,0.55955
|
||||
3579,622,0.10045,0.70594
|
||||
3580,624,0.090376,0.28564
|
||||
3581,625,0.055574,0.24113
|
||||
3582,626,0.060658,0.30453
|
||||
3583,627,0.088106,0.42489
|
||||
3584,628,0.10668,0.57663
|
||||
3585,629,0.11436,0.73438
|
||||
3586,631,0.092673,0.27996
|
||||
3587,632,0.070837,0.24312
|
||||
3588,633,0.071837,0.31331
|
||||
3589,634,0.10559,0.45099
|
||||
3590,635,0.055948,0.61128
|
||||
3591,636,0.10764,0.78426
|
||||
3592,638,0.099881,0.28908
|
||||
3593,639,0.058087,0.26122
|
||||
3594,640,0.049617,0.3359
|
||||
3595,641,0.033961,0.4745
|
||||
3596,642,0.076609,0.6363
|
||||
3597,644,0.077513,0.39055
|
||||
3598,645,0.052841,0.27914
|
||||
3599,646,0.080118,0.27985
|
||||
3600,647,0.06745,0.40313
|
||||
3601,648,0.068639,0.57203
|
||||
3602,650,0.10875,0.381
|
||||
3603,651,0.064039,0.23013
|
||||
3604,652,0.06631,0.25651
|
||||
3605,653,0.06431,0.43451
|
||||
3606,655,0.089584,0.43654
|
||||
3607,656,0.095025,0.2244
|
||||
3608,657,0.046494,0.16451
|
||||
3609,658,0.062422,0.34744
|
||||
3610,660,0.065911,0.47125
|
||||
3611,661,0.087185,0.23454
|
||||
3612,662,0.043045,0.094604
|
||||
3613,663,0.051207,0.29061
|
||||
3614,664,0.07003,0.53155
|
||||
3615,666,0.058302,0.27346
|
||||
3616,667,0.044426,0.075985
|
||||
3617,668,0.060829,0.22237
|
||||
3618,669,0.082533,0.44624
|
||||
3619,671,0.095735,0.3995
|
||||
3620,672,0.074451,0.19306
|
||||
3621,673,0.03985,0.04715
|
||||
3622,674,0.065908,0.22087
|
||||
3623,675,0.052044,0.40935
|
||||
3624,677,0.068861,0.48659
|
||||
3625,677,0.065753,0.78468
|
||||
3626,679,0.090192,0.12168
|
||||
3627,680,0.044919,0.089761
|
||||
3628,681,0.05526,0.26014
|
||||
3629,682,0.090563,0.44568
|
||||
3630,683,0.083276,0.62779
|
||||
3631,685,0.090358,0.29193
|
||||
3632,686,0.047125,0.15728
|
||||
3633,687,0.06156,0.17626
|
||||
3634,688,0.059814,0.31989
|
||||
3635,690,0.087951,0.63475
|
||||
3636,690,0.088435,0.6424
|
||||
3637,692,0.064857,0.32143
|
||||
3638,693,0.061375,0.22204
|
||||
3639,694,0.03735,0.24184
|
||||
3640,695,0.076527,0.34438
|
||||
3641,696,0.071587,0.46863
|
||||
3642,697,0.092114,0.59412
|
||||
3643,699,0.043643,0.38307
|
||||
3644,700,0.050312,0.2936
|
||||
3645,701,0.060029,0.25528
|
||||
3646,702,0.049366,0.29442
|
||||
3647,703,0.064904,0.38212
|
||||
3648,704,0.072242,0.49863
|
||||
3649,706,0.075988,0.45218
|
||||
3650,707,0.0769,0.37363
|
||||
3651,708,0.072505,0.34137
|
||||
3652,709,0.07811,0.36967
|
||||
3653,710,0.078036,0.44525
|
||||
3654,712,0.073206,0.49596
|
||||
3655,713,0.068118,0.41128
|
||||
3656,714,0.050477,0.3862
|
||||
3657,715,0.064732,0.44054
|
||||
3658,717,0.076224,0.48555
|
||||
3659,717,0.084693,0.67737
|
||||
3660,719,0.078924,0.45158
|
||||
3661,720,0.10023,0.53984
|
||||
3662,721,0.10665,0.66459
|
||||
3663,723,0.096093,0.51577
|
||||
3664,724,0.14278,0.62171
|
||||
3665,725,0.14953,0.74938
|
||||
3666,728,0.13766,0.70935
|
||||
3667,728,0.13118,0.76398
|
||||
3668,729,0.13124,0.88244
|
||||
3669,732,0.15585,1.0328
|
||||
3670,733,0.17714,1.125
|
||||
3679,741,0.19688,1.8353
|
||||
3689,749,0.17713,1.7566
|
||||
3690,749,0.17868,1.7155
|
||||
3691,751,0.1978,1.595
|
||||
3692,752,0.16191,1.5302
|
||||
3693,753,0.15975,1.4976
|
||||
3694,754,0.18196,1.4574
|
||||
3695,755,0.17917,1.4232
|
||||
3696,756,0.15793,1.376
|
||||
3697,756,0.15715,1.289
|
||||
3698,757,0.18161,1.2367
|
||||
3699,758,0.19893,1.2009
|
||||
3701,761,0.19814,1.1338
|
||||
3702,762,0.19765,1.0772
|
||||
3704,764,0.15639,0.96618
|
||||
3705,765,0.14924,0.91483
|
||||
3706,767,0.16049,1.1076
|
||||
3707,768,0.14661,1.0304
|
||||
3708,769,0.16173,0.95107
|
||||
3709,770,0.17248,0.88029
|
||||
3710,771,0.15391,0.80607
|
||||
3711,772,0.12899,0.74643
|
||||
3712,773,0.14391,0.69723
|
||||
3713,774,0.10839,0.66246
|
||||
3714,775,0.13295,0.62635
|
||||
3715,777,0.11932,0.99003
|
||||
3716,777,0.11136,0.60216
|
||||
3717,778,0.11118,0.61244
|
||||
3718,780,0.084911,0.75093
|
||||
3719,781,0.084824,0.65949
|
||||
3720,782,0.093516,0.57417
|
||||
3721,783,0.083243,0.49306
|
||||
3722,784,0.075382,0.42461
|
||||
3723,786,0.08921,0.97408
|
||||
3724,787,0.063303,0.83625
|
||||
3725,788,0.055663,0.69642
|
||||
3726,789,0.032825,0.55779
|
||||
3727,790,0.033788,0.42876
|
||||
3728,791,0.056153,0.33518
|
||||
3729,792,0.077125,0.32126
|
||||
3730,794,0.098905,0.74103
|
||||
3731,795,0.093547,0.58313
|
||||
3732,796,0.072227,0.44562
|
||||
3733,797,0.069637,0.35809
|
||||
3734,798,0.072181,0.36877
|
||||
3735,800,0.1012,0.73319
|
||||
3736,801,0.088988,0.59052
|
||||
3737,802,0.081669,0.49135
|
||||
3738,803,0.088109,0.45904
|
||||
3739,805,0.14143,0.87342
|
||||
3740,806,0.088275,0.72476
|
||||
3741,807,0.081838,0.60473
|
||||
3742,808,0.061756,0.5184
|
||||
3743,809,0.07685,0.49552
|
||||
3744,810,0.083131,0.54533
|
||||
3745,812,0.07936,0.65237
|
||||
3746,813,0.11602,0.52991
|
||||
3747,814,0.092008,0.46097
|
||||
3748,815,0.11438,0.46278
|
||||
3749,817,0.11669,0.69024
|
||||
3750,818,0.12787,0.54126
|
||||
3751,819,0.1183,0.4249
|
||||
3752,820,0.11498,0.36484
|
||||
3753,821,0.12448,0.3726
|
||||
3754,822,0.12017,0.43852
|
||||
3755,824,0.075637,0.46155
|
||||
3756,825,0.067569,0.33571
|
||||
3757,826,0.086052,0.26541
|
||||
3758,828,0.054262,0.62002
|
||||
3759,829,0.059315,0.43646
|
||||
3760,830,0.054124,0.28347
|
||||
3761,832,0.045085,0.61395
|
||||
3762,833,0.039748,0.38973
|
||||
3763,835,0.054577,0.68335
|
||||
3764,836,0.034231,0.44815
|
||||
3765,837,0.051626,0.29632
|
||||
3766,839,0.048617,0.49034
|
||||
3767,840,0.035521,0.32485
|
||||
3768,842,0.02213,0.51578
|
||||
3769,843,0.038473,0.3297
|
||||
3770,845,0.03616,0.5392
|
||||
3771,846,0.069148,0.3313
|
||||
3772,848,0.058476,0.58235
|
||||
3773,849,0.033742,0.36661
|
||||
3774,850,0.068282,0.27062
|
||||
3775,852,0.055623,0.43044
|
||||
3776,853,0.081876,0.28355
|
||||
3777,855,0.085668,0.57836
|
||||
3778,856,0.079642,0.39534
|
||||
3779,857,0.077434,0.30271
|
||||
3780,859,0.075026,0.5724
|
||||
3781,860,0.052223,0.38786
|
||||
3782,861,0.083688,0.32685
|
||||
3783,863,0.093958,0.66109
|
||||
3784,864,0.071433,0.48336
|
||||
3785,865,0.097187,0.38096
|
||||
3786,867,0.081609,0.7876
|
||||
3787,868,0.064721,0.63527
|
||||
3788,869,0.043929,0.50521
|
||||
3789,870,0.047902,0.43407
|
||||
3790,872,0.07444,0.84348
|
||||
3791,873,0.061356,0.67386
|
||||
3792,874,0.051737,0.54037
|
||||
3793,875,0.050476,0.44854
|
||||
3794,876,0.068758,0.44399
|
||||
3795,878,0.10942,0.80398
|
||||
3796,879,0.05869,0.64458
|
||||
3797,880,0.04027,0.52199
|
||||
3798,881,0.013119,0.45485
|
||||
3799,882,0.066674,0.46733
|
||||
3800,884,0.078149,0.85938
|
||||
3801,885,0.075164,0.72029
|
||||
3802,886,0.068032,0.60417
|
||||
3803,887,0.056921,0.52924
|
||||
3804,888,0.032784,0.51512
|
||||
3805,889,0.068174,0.55505
|
||||
3806,891,0.10477,0.80946
|
||||
3807,892,0.095494,0.67578
|
||||
3808,893,0.062951,0.57035
|
||||
3809,894,0.062536,0.51395
|
||||
3810,895,0.072229,0.51577
|
||||
3811,896,0.095976,0.58189
|
||||
3812,898,0.07504,0.64844
|
||||
3813,899,0.079638,0.52944
|
||||
3814,900,0.026548,0.45961
|
||||
3815,901,0.096918,0.4666
|
||||
3816,902,0.13836,0.53983
|
||||
3817,903,0.10539,0.66673
|
||||
3818,905,0.099527,0.54633
|
||||
3819,906,0.060867,0.48184
|
||||
3820,907,0.074188,0.49242
|
||||
3821,908,0.094226,0.56528
|
||||
3822,910,0.08159,0.66476
|
||||
3823,911,0.074229,0.54405
|
||||
3824,912,0.081451,0.46848
|
||||
3825,913,0.072537,0.46444
|
||||
3826,914,0.13248,0.5335
|
||||
3827,916,0.1075,0.56961
|
||||
3828,917,0.089515,0.42794
|
||||
3829,918,0.10344,0.35664
|
||||
3830,919,0.11292,0.39764
|
||||
3831,921,0.11373,0.5034
|
||||
3832,922,0.11061,0.31533
|
||||
3833,923,0.12234,0.25132
|
||||
3834,925,0.095064,0.46486
|
||||
3835,926,0.11477,0.2108
|
||||
3836,928,0.1148,0.55532
|
||||
3837,929,0.13395,0.19798
|
||||
3838,931,0.089244,0.42762
|
||||
3839,932,0.10884,0.061937
|
||||
3840,934,0.09651,0.16994
|
||||
3841,936,0.11766,0.27467
|
||||
3842,939,0.15204,0.86704
|
||||
3843,940,0.10821,0.35631
|
||||
3844,943,0.10591,0.92646
|
||||
3845,945,0.11257,1.0528
|
||||
3846,947,0.14007,1.2791
|
||||
4086,4026,0.19647,71.785
|
||||
4087,4029,0.17798,69.611
|
||||
4098,4039,0.1933,71.937
|
||||
4104,4032,0.19953,87.231
|
||||
4107,4035,0.18493,87.513
|
||||
4108,4035,0.16613,88.736
|
||||
4109,4022,0.18817,104.65
|
||||
4113,4056,0.19922,70.563
|
||||
4114,4027,0.19434,105.02
|
||||
4117,4058,0.19995,73.055
|
||||
4119,4058,0.17109,75.484
|
||||
4120,4059,0.17764,75.503
|
||||
4122,4063,0.18719,73.119
|
||||
4155,4089,0.17875,80.876
|
||||
4156,4090,0.18251,80.874
|
||||
4167,4104,0.19689,77.014
|
||||
4168,4081,0.1964,106.63
|
||||
4174,4054,0.19218,146.25
|
||||
4175,4053,0.18144,148.57
|
||||
4176,4053,0.1844,149.74
|
||||
4177,4057,0.18942,146.28
|
||||
4178,4056,0.19632,148.61
|
||||
4179,4058,0.17388,147.46
|
||||
4180,4058,0.14882,148.64
|
||||
4181,4058,0.15282,149.81
|
||||
4182,4058,0.16478,150.98
|
||||
4183,4058,0.16937,152.15
|
||||
4184,4063,0.16278,147.48
|
||||
4185,4060,0.16456,152.18
|
||||
4186,4062,0.17425,151.01
|
||||
4188,4063,0.173,152.2
|
||||
4189,4090,0.18209,120.6
|
||||
4192,4068,0.1988,151.06
|
||||
4212,4042,0.19878,204.79
|
||||
4213,4042,0.19257,206.02
|
||||
4346,4232,0.19848,147.54
|
||||
4449,1,0.13053,1.2687
|
||||
4450,1,0.065646,1.6334
|
||||
4451,2,0.11856,1.3391
|
||||
4452,2,0.12076,1.885
|
||||
4453,4,0.07551,0.85034
|
||||
4454,5,0.069256,0.65579
|
||||
4455,6,0.12955,0.48
|
||||
4456,6,0.14076,1.1592
|
||||
4457,8,0.1402,0.20551
|
||||
4458,8,0.12864,0.92316
|
||||
4459,9,0.1212,0.83523
|
||||
4460,10,0.091014,0.76869
|
||||
4461,11,0.094309,0.71579
|
||||
4462,12,0.09095,0.68183
|
||||
4463,13,0.086022,0.66145
|
||||
4464,14,0.12145,0.66037
|
||||
4465,15,0.11576,0.67138
|
||||
4466,16,0.10502,0.6999
|
||||
4467,18,0.13693,0.16018
|
||||
4468,19,0.12991,0.14666
|
||||
4469,19,0.1361,0.83053
|
||||
4470,21,0.11473,0.13735
|
||||
4471,22,0.12905,0.1519
|
||||
4472,23,0.11426,0.17233
|
||||
4473,24,0.11572,0.2005
|
||||
4474,25,0.094085,0.24349
|
||||
4475,26,0.11117,0.2909
|
||||
4476,27,0.069105,0.35066
|
||||
4477,28,0.12759,0.42191
|
||||
4478,29,0.12165,0.50369
|
||||
4479,30,0.12076,0.59271
|
||||
4480,31,0.1146,0.68602
|
||||
4481,32,0.11854,0.78384
|
||||
4482,34,0.10691,0.11035
|
||||
4483,35,0.095109,0.04609
|
||||
4484,36,0.1005,0.12729
|
||||
4485,37,0.082428,0.23604
|
||||
4486,38,0.10448,0.33445
|
||||
4487,39,0.080922,0.433
|
||||
4488,40,0.11181,0.51503
|
||||
4489,41,0.10755,0.55844
|
||||
4490,43,0.14419,0.45751
|
||||
4491,44,0.091988,0.38999
|
||||
4492,45,0.068163,0.32214
|
||||
4493,46,0.096962,0.24878
|
||||
4494,47,0.076357,0.19398
|
||||
4495,48,0.067854,0.17365
|
||||
4496,49,0.081921,0.21149
|
||||
4497,50,0.11233,0.29649
|
||||
4498,52,0.079512,0.64139
|
||||
4499,53,0.074202,0.49544
|
||||
4500,54,0.055416,0.35099
|
||||
4501,55,0.04999,0.20826
|
||||
4502,56,0.076014,0.096245
|
||||
4503,57,0.076982,0.19117
|
||||
4504,59,0.10069,0.60803
|
||||
4505,60,0.066785,0.41546
|
||||
4506,61,0.060471,0.21604
|
||||
4507,62,0.078096,0.010787
|
||||
4508,64,0.11174,0.74511
|
||||
4509,65,0.072687,0.51088
|
||||
4510,66,0.085603,0.26699
|
||||
4511,67,0.068949,0.066393
|
||||
4512,69,0.084442,0.63513
|
||||
4513,70,0.085138,0.33661
|
||||
4514,71,0.076727,0.12166
|
||||
4515,73,0.076458,0.5219
|
||||
4516,75,0.091597,0.97262
|
||||
4517,76,0.11259,0.61147
|
||||
4518,78,0.11267,0.98103
|
||||
4519,79,0.10949,0.58178
|
||||
4520,81,0.085893,0.82734
|
||||
4521,83,0.090288,1.028
|
||||
4522,84,0.14685,0.59582
|
||||
4523,87,0.13908,1.1813
|
||||
4524,89,0.10489,1.1831
|
||||
4525,91,0.068849,1.1217
|
||||
4526,92,0.098152,0.61523
|
||||
4527,96,0.13071,1.2383
|
||||
4528,97,0.078189,0.58424
|
||||
4529,100,0.052122,0.6118
|
||||
4530,102,0.095975,0.30614
|
||||
4531,104,0.17613,0.50631
|
||||
4536,1557,0.11952,2.3321
|
||||
4537,1555,0.090415,2.0274
|
||||
4538,1555,0.16563,0.95465
|
||||
|
Binary file not shown.
|
Before Width: | Height: | Size: 61 KiB |
File diff suppressed because it is too large
Load Diff
|
|
@ -1,261 +0,0 @@
|
|||
% function lineStyles = linspecer(N)
|
||||
% This function creates an Nx3 array of N [R B G] colors
|
||||
% These can be used to plot lots of lines with distinguishable and nice
|
||||
% looking colors.
|
||||
%
|
||||
% lineStyles = linspecer(N); makes N colors for you to use: lineStyles(ii,:)
|
||||
%
|
||||
% colormap(linspecer); set your colormap to have easily distinguishable
|
||||
% colors and a pleasing aesthetic
|
||||
%
|
||||
% lineStyles = linspecer(N,'qualitative'); forces the colors to all be distinguishable (up to 12)
|
||||
% lineStyles = linspecer(N,'sequential'); forces the colors to vary along a spectrum
|
||||
%
|
||||
% % Examples demonstrating the colors.
|
||||
%
|
||||
% LINE COLORS
|
||||
% N=6;
|
||||
% X = linspace(0,pi*3,1000);
|
||||
% Y = bsxfun(@(x,n)sin(x+2*n*pi/N), X.', 1:N);
|
||||
% C = linspecer(N);
|
||||
% axes('NextPlot','replacechildren', 'ColorOrder',C);
|
||||
% plot(X,Y,'linewidth',5)
|
||||
% ylim([-1.1 1.1]);
|
||||
%
|
||||
% SIMPLER LINE COLOR EXAMPLE
|
||||
% N = 6; X = linspace(0,pi*3,1000);
|
||||
% C = linspecer(N)
|
||||
% hold off;
|
||||
% for ii=1:N
|
||||
% Y = sin(X+2*ii*pi/N);
|
||||
% plot(X,Y,'color',C(ii,:),'linewidth',3);
|
||||
% hold on;
|
||||
% end
|
||||
%
|
||||
% COLORMAP EXAMPLE
|
||||
% A = rand(15);
|
||||
% figure; imagesc(A); % default colormap
|
||||
% figure; imagesc(A); colormap(linspecer); % linspecer colormap
|
||||
%
|
||||
% See also NDHIST, NHIST, PLOT, COLORMAP, 43700-cubehelix-colormaps
|
||||
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
|
||||
% by Jonathan Lansey, March 2009-2013 – Lansey at gmail.com %
|
||||
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
|
||||
%
|
||||
%% credits and where the function came from
|
||||
% The colors are largely taken from:
|
||||
% http://colorbrewer2.org and Cynthia Brewer, Mark Harrower and The Pennsylvania State University
|
||||
%
|
||||
%
|
||||
% She studied this from a phsychometric perspective and crafted the colors
|
||||
% beautifully.
|
||||
%
|
||||
% I made choices from the many there to decide the nicest once for plotting
|
||||
% lines in Matlab. I also made a small change to one of the colors I
|
||||
% thought was a bit too bright. In addition some interpolation is going on
|
||||
% for the sequential line styles.
|
||||
%
|
||||
%
|
||||
%%
|
||||
|
||||
function lineStyles=linspecer(N,varargin)
|
||||
|
||||
if nargin==0 % return a colormap
|
||||
lineStyles = linspecer(128);
|
||||
return;
|
||||
end
|
||||
|
||||
if ischar(N)
|
||||
lineStyles = linspecer(128,N);
|
||||
return;
|
||||
end
|
||||
|
||||
if N<=0 % its empty, nothing else to do here
|
||||
lineStyles=[];
|
||||
return;
|
||||
end
|
||||
|
||||
% interperet varagin
|
||||
qualFlag = 0;
|
||||
colorblindFlag = 0;
|
||||
|
||||
if ~isempty(varargin)>0 % you set a parameter?
|
||||
switch lower(varargin{1})
|
||||
case {'qualitative','qua'}
|
||||
if N>12 % go home, you just can't get this.
|
||||
warning('qualitiative is not possible for greater than 12 items, please reconsider');
|
||||
else
|
||||
if N>9
|
||||
warning(['Default may be nicer for ' num2str(N) ' for clearer colors use: whitebg(''black''); ']);
|
||||
end
|
||||
end
|
||||
qualFlag = 1;
|
||||
case {'sequential','seq'}
|
||||
lineStyles = colorm(N);
|
||||
return;
|
||||
case {'white','whitefade'}
|
||||
lineStyles = whiteFade(N);return;
|
||||
case 'red'
|
||||
lineStyles = whiteFade(N,'red');return;
|
||||
case 'blue'
|
||||
lineStyles = whiteFade(N,'blue');return;
|
||||
case 'green'
|
||||
lineStyles = whiteFade(N,'green');return;
|
||||
case {'gray','grey'}
|
||||
lineStyles = whiteFade(N,'gray');return;
|
||||
case {'colorblind'}
|
||||
colorblindFlag = 1;
|
||||
otherwise
|
||||
warning(['parameter ''' varargin{1} ''' not recognized']);
|
||||
end
|
||||
end
|
||||
% *.95
|
||||
% predefine some colormaps
|
||||
set3 = colorBrew2mat({[141, 211, 199];[ 255, 237, 111];[ 190, 186, 218];[ 251, 128, 114];[ 128, 177, 211];[ 253, 180, 98];[ 179, 222, 105];[ 188, 128, 189];[ 217, 217, 217];[ 204, 235, 197];[ 252, 205, 229];[ 255, 255, 179]}');
|
||||
set1JL = brighten(colorBrew2mat({[228, 26, 28];[ 55, 126, 184]; [ 77, 175, 74];[ 255, 127, 0];[ 255, 237, 111]*.85;[ 166, 86, 40];[ 247, 129, 191];[ 153, 153, 153];[ 152, 78, 163]}'));
|
||||
set1 = brighten(colorBrew2mat({[ 55, 126, 184]*.85;[228, 26, 28];[ 77, 175, 74];[ 255, 127, 0];[ 152, 78, 163]}),.8);
|
||||
|
||||
% colorblindSet = {[215,25,28];[253,174,97];[171,217,233];[44,123,182]};
|
||||
colorblindSet = {[215,25,28];[253,174,97];[171,217,233]*.8;[44,123,182]*.8};
|
||||
|
||||
set3 = dim(set3,.93);
|
||||
|
||||
if colorblindFlag
|
||||
switch N
|
||||
% sorry about this line folks. kind of legacy here because I used to
|
||||
% use individual 1x3 cells instead of nx3 arrays
|
||||
case 4
|
||||
lineStyles = colorBrew2mat(colorblindSet);
|
||||
otherwise
|
||||
colorblindFlag = false;
|
||||
warning('sorry unsupported colorblind set for this number, using regular types');
|
||||
end
|
||||
end
|
||||
if ~colorblindFlag
|
||||
switch N
|
||||
case 1
|
||||
lineStyles = { [ 55, 126, 184]/255};
|
||||
case {2, 3, 4, 5 }
|
||||
lineStyles = set1(1:N);
|
||||
case {6 , 7, 8, 9}
|
||||
lineStyles = set1JL(1:N)';
|
||||
case {10, 11, 12}
|
||||
if qualFlag % force qualitative graphs
|
||||
lineStyles = set3(1:N)';
|
||||
else % 10 is a good number to start with the sequential ones.
|
||||
lineStyles = cmap2linspecer(colorm(N));
|
||||
end
|
||||
otherwise % any old case where I need a quick job done.
|
||||
lineStyles = cmap2linspecer(colorm(N));
|
||||
end
|
||||
end
|
||||
lineStyles = cell2mat(lineStyles);
|
||||
|
||||
end
|
||||
|
||||
% extra functions
|
||||
function varIn = colorBrew2mat(varIn)
|
||||
for ii=1:length(varIn) % just divide by 255
|
||||
varIn{ii}=varIn{ii}/255;
|
||||
end
|
||||
end
|
||||
|
||||
function varIn = brighten(varIn,varargin) % increase the brightness
|
||||
|
||||
if isempty(varargin),
|
||||
frac = .9;
|
||||
else
|
||||
frac = varargin{1};
|
||||
end
|
||||
|
||||
for ii=1:length(varIn)
|
||||
varIn{ii}=varIn{ii}*frac+(1-frac);
|
||||
end
|
||||
end
|
||||
|
||||
function varIn = dim(varIn,f)
|
||||
for ii=1:length(varIn)
|
||||
varIn{ii} = f*varIn{ii};
|
||||
end
|
||||
end
|
||||
|
||||
function vOut = cmap2linspecer(vIn) % changes the format from a double array to a cell array with the right format
|
||||
vOut = cell(size(vIn,1),1);
|
||||
for ii=1:size(vIn,1)
|
||||
vOut{ii} = vIn(ii,:);
|
||||
end
|
||||
end
|
||||
%%
|
||||
% colorm returns a colormap which is really good for creating informative
|
||||
% heatmap style figures.
|
||||
% No particular color stands out and it doesn't do too badly for colorblind people either.
|
||||
% It works by interpolating the data from the
|
||||
% 'spectral' setting on http://colorbrewer2.org/ set to 11 colors
|
||||
% It is modified a little to make the brightest yellow a little less bright.
|
||||
function cmap = colorm(varargin)
|
||||
n = 100;
|
||||
if ~isempty(varargin)
|
||||
n = varargin{1};
|
||||
end
|
||||
|
||||
if n==1
|
||||
cmap = [0.2005 0.5593 0.7380];
|
||||
return;
|
||||
end
|
||||
if n==2
|
||||
cmap = [0.2005 0.5593 0.7380;
|
||||
0.9684 0.4799 0.2723];
|
||||
return;
|
||||
end
|
||||
|
||||
frac=.95; % Slight modification from colorbrewer here to make the yellows in the center just a bit darker
|
||||
cmapp = [158, 1, 66; 213, 62, 79; 244, 109, 67; 253, 174, 97; 254, 224, 139; 255*frac, 255*frac, 191*frac; 230, 245, 152; 171, 221, 164; 102, 194, 165; 50, 136, 189; 94, 79, 162];
|
||||
x = linspace(1,n,size(cmapp,1));
|
||||
xi = 1:n;
|
||||
cmap = zeros(n,3);
|
||||
for ii=1:3
|
||||
cmap(:,ii) = pchip(x,cmapp(:,ii),xi);
|
||||
end
|
||||
cmap = flipud(cmap/255);
|
||||
end
|
||||
|
||||
function cmap = whiteFade(varargin)
|
||||
n = 100;
|
||||
if nargin>0
|
||||
n = varargin{1};
|
||||
end
|
||||
|
||||
thisColor = 'blue';
|
||||
|
||||
if nargin>1
|
||||
thisColor = varargin{2};
|
||||
end
|
||||
switch thisColor
|
||||
case {'gray','grey'}
|
||||
cmapp = [255,255,255;240,240,240;217,217,217;189,189,189;150,150,150;115,115,115;82,82,82;37,37,37;0,0,0];
|
||||
case 'green'
|
||||
cmapp = [247,252,245;229,245,224;199,233,192;161,217,155;116,196,118;65,171,93;35,139,69;0,109,44;0,68,27];
|
||||
case 'blue'
|
||||
cmapp = [247,251,255;222,235,247;198,219,239;158,202,225;107,174,214;66,146,198;33,113,181;8,81,156;8,48,107];
|
||||
case 'red'
|
||||
cmapp = [255,245,240;254,224,210;252,187,161;252,146,114;251,106,74;239,59,44;203,24,29;165,15,21;103,0,13];
|
||||
otherwise
|
||||
warning(['sorry your color argument ' thisColor ' was not recognized']);
|
||||
end
|
||||
|
||||
cmap = interpomap(n,cmapp);
|
||||
end
|
||||
|
||||
% Eat a approximate colormap, then interpolate the rest of it up.
|
||||
function cmap = interpomap(n,cmapp)
|
||||
x = linspace(1,n,size(cmapp,1));
|
||||
xi = 1:n;
|
||||
cmap = zeros(n,3);
|
||||
for ii=1:3
|
||||
cmap(:,ii) = pchip(x,cmapp(:,ii),xi);
|
||||
end
|
||||
cmap = (cmap/255); % flipud??
|
||||
end
|
||||
|
||||
|
||||
|
||||
|
|
@ -1,154 +0,0 @@
|
|||
clear; clc;
|
||||
|
||||
addpath(genpath('src'));
|
||||
addpath(genpath('data'));
|
||||
|
||||
%% data preparation
|
||||
global data_path;
|
||||
% your directory should contain files like this
|
||||
% - 00
|
||||
% l- 00.csv (gt pose)
|
||||
% l- velodyne
|
||||
% l- <0xxxx.bin>
|
||||
data_path = '/media/gskim/Data/KITTI odo/data_odometry_velodyne/dataset/sequences/00/';
|
||||
|
||||
|
||||
down_shape = [40, 120];
|
||||
skip_data_frame = 1;
|
||||
[data_scancontexts, data_ringkeys, data_poses] = loadData(down_shape, skip_data_frame);
|
||||
|
||||
figure(101); clf;
|
||||
plot(data_poses(:,1), data_poses(:,2));
|
||||
axis equal;
|
||||
|
||||
%% main - global recognizer
|
||||
revisit_criteria = 5; % in meter (recommend test for 5, 10, 20 meters)
|
||||
keyframe_gap = 1; % for_fast_eval (if 1, no skip)
|
||||
|
||||
global num_candidates; num_candidates = 50;
|
||||
% global num_node_enough_apart; num_node_enough_apart = 50;
|
||||
|
||||
% policy (top N)
|
||||
num_top_n = 25;
|
||||
top_n = linspace(1, num_top_n, num_top_n);
|
||||
|
||||
% Entropy thresholds
|
||||
middle_thres = 0.01;
|
||||
thresholds1 = linspace(0, middle_thres, 50);
|
||||
thresholds2 = linspace(middle_thres, 1, 50);
|
||||
thresholds = [thresholds1, thresholds2];
|
||||
num_thresholds = length(thresholds);
|
||||
|
||||
% Main variables to store the result for drawing PR curve
|
||||
num_hits = zeros(num_top_n, num_thresholds);
|
||||
num_false_alarms = zeros(num_top_n, num_thresholds);
|
||||
num_correct_rejections = zeros(num_top_n, num_thresholds);
|
||||
num_misses = zeros(num_top_n, num_thresholds);
|
||||
|
||||
% main
|
||||
loop_log = [];
|
||||
|
||||
exp_poses = [];
|
||||
exp_ringkeys = [];
|
||||
exp_scancontexts = {};
|
||||
|
||||
num_queries = length(data_poses);
|
||||
for query_idx = 1:num_queries - 1
|
||||
|
||||
% save to (online) DB
|
||||
query_sc = data_scancontexts{query_idx};
|
||||
query_rk = data_ringkeys(query_idx, :);
|
||||
query_pose = data_poses(query_idx,:);
|
||||
|
||||
exp_scancontexts{end+1} = query_sc;
|
||||
exp_poses = [exp_poses; query_pose];
|
||||
exp_ringkeys = [exp_ringkeys; query_rk];
|
||||
|
||||
if(rem(query_idx, keyframe_gap) ~= 0)
|
||||
continue;
|
||||
end
|
||||
|
||||
if( length(exp_scancontexts) < num_candidates )
|
||||
continue;
|
||||
end
|
||||
|
||||
tree = createns(exp_ringkeys(1:end-(num_candidates-1), :), 'NSMethod', 'kdtree'); % Create object to use in k-nearest neighbor search
|
||||
|
||||
% revisitness
|
||||
[revisitness, how_far_apart] = isRevisitGlobalLoc(query_pose, exp_poses(1:end-(num_candidates-1), :), revisit_criteria);
|
||||
disp([revisitness, how_far_apart])
|
||||
|
||||
% find candidates
|
||||
candidates = knnsearch(tree, query_rk, 'K', num_candidates);
|
||||
|
||||
% find the nearest (top 1) via pairwise comparison
|
||||
nearest_idx = 0;
|
||||
min_dist = inf; % initialization
|
||||
for ith_candidate = 1:length(candidates)
|
||||
candidate_node_idx = candidates(ith_candidate);
|
||||
candidate_img = exp_scancontexts{candidate_node_idx};
|
||||
|
||||
distance_to_query = sc_dist(query_sc, candidate_img);
|
||||
|
||||
if( distance_to_query < min_dist)
|
||||
nearest_idx = candidate_node_idx;
|
||||
min_dist = distance_to_query;
|
||||
end
|
||||
end
|
||||
|
||||
% prcurve analysis
|
||||
for topk = 1:num_top_n
|
||||
for thres_idx = 1:num_thresholds
|
||||
threshold = thresholds(thres_idx);
|
||||
|
||||
reject = 0;
|
||||
if( min_dist > threshold)
|
||||
reject = 1;
|
||||
end
|
||||
|
||||
if(reject == 1)
|
||||
if(revisitness == 0)
|
||||
% TN: Correct Rejection
|
||||
num_correct_rejections(topk, thres_idx) = num_correct_rejections(topk, thres_idx) + 1;
|
||||
else
|
||||
% FN: MISS
|
||||
num_misses(topk, thres_idx) = num_misses(topk, thres_idx) + 1;
|
||||
end
|
||||
else
|
||||
% if under the theshold, it is considered seen.
|
||||
% and then check the correctness
|
||||
if( dist_btn_pose(query_pose, exp_poses(nearest_idx, :)) < revisit_criteria)
|
||||
% TP: Hit
|
||||
num_hits(topk, thres_idx) = num_hits(topk, thres_idx) + 1;
|
||||
else
|
||||
% FP: False Alarm
|
||||
num_false_alarms(topk, thres_idx) = num_false_alarms(topk, thres_idx) + 1;
|
||||
end
|
||||
end
|
||||
|
||||
end
|
||||
end
|
||||
|
||||
if( rem(query_idx, 100) == 0)
|
||||
disp( strcat(num2str(query_idx/num_queries * 100), ' % processed') );
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
||||
%% save the log
|
||||
savePath = strcat("pr_result/within ", num2str(revisit_criteria), "m/");
|
||||
if((~7==exist(savePath,'dir')))
|
||||
mkdir(savePath);
|
||||
end
|
||||
save(strcat(savePath, 'nCorrectRejections.mat'), 'num_correct_rejections');
|
||||
save(strcat(savePath, 'nMisses.mat'), 'num_misses');
|
||||
save(strcat(savePath, 'nHits.mat'), 'num_hits');
|
||||
save(strcat(savePath, 'nFalseAlarms.mat'), 'num_false_alarms');
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
|
@ -1,130 +0,0 @@
|
|||
ResultsDir = './pr_result/';
|
||||
|
||||
%%
|
||||
title_str = strcat('KITTI 00');
|
||||
|
||||
%% Params
|
||||
FigIdx = 2;
|
||||
figure(FigIdx); clf;
|
||||
|
||||
TopNindexes = [1];
|
||||
name = 'top1';
|
||||
|
||||
nTopNindexes = length(TopNindexes);
|
||||
|
||||
%% Main
|
||||
SequenceNames = dir(ResultsDir); SequenceNames(1:2, :) = []; SequenceNames = {SequenceNames(:).name};
|
||||
nSequences = length(SequenceNames);
|
||||
|
||||
all_Precisions = {};
|
||||
all_Recalls = {};
|
||||
|
||||
for ithTopN = 1:nTopNindexes
|
||||
|
||||
TopNidx = TopNindexes(ithTopN);
|
||||
|
||||
line_width = 4;
|
||||
|
||||
LineColors = colorcube(nSequences);
|
||||
LineColors = linspecer(nSequences,'qualitative');
|
||||
% LineColors = linspecer(nSequences,'sequential');
|
||||
LineColors = flipud(LineColors);
|
||||
|
||||
AUCs = zeros(1, nSequences);
|
||||
for ithSeq = 1:nSequences
|
||||
|
||||
% seq info
|
||||
ithSeqName = SequenceNames{ithSeq};
|
||||
SequenceNames{ithSeq} = string(ithSeqName);
|
||||
|
||||
ithSeqPath = strcat(ResultsDir, ithSeqName, '/');
|
||||
ithSeqPRcurveData = dir(ithSeqPath); ithSeqPRcurveData(1:2, :) = []; ithSeqPRcurveData = {ithSeqPRcurveData(:).name};
|
||||
|
||||
% load
|
||||
nCorrectRejectionsAll = load(strcat(ithSeqPath, ithSeqPRcurveData{1}));
|
||||
nCorrectRejectionsAll = nCorrectRejectionsAll.num_correct_rejections;
|
||||
nCorrectRejectionsForThisTopN = nCorrectRejectionsAll(TopNidx, :);
|
||||
|
||||
nFalseAlarmsAll = load(strcat(ithSeqPath, ithSeqPRcurveData{2}));
|
||||
nFalseAlarmsAll = nFalseAlarmsAll.num_false_alarms;
|
||||
nFalseAlarmsForThisTopN = nFalseAlarmsAll(TopNidx, :);
|
||||
|
||||
nHitsAll = load(strcat(ithSeqPath, ithSeqPRcurveData{3}));
|
||||
nHitsAll = nHitsAll.num_hits;
|
||||
nHitsForThisTopN = nHitsAll(TopNidx, :);
|
||||
|
||||
nMissesAll = load(strcat(ithSeqPath, ithSeqPRcurveData{4}));
|
||||
nMissesAll = nMissesAll.num_misses;
|
||||
nMissesForThisTopN = nMissesAll(TopNidx, :);
|
||||
|
||||
% info
|
||||
nTopNs = size(nCorrectRejectionsAll, 1);
|
||||
nThres = size(nCorrectRejectionsAll, 2);
|
||||
|
||||
% main
|
||||
Precisions = [];
|
||||
Recalls = [];
|
||||
Accuracies = [];
|
||||
for ithThres = 1:nThres
|
||||
nCorrectRejections = nCorrectRejectionsForThisTopN(ithThres);
|
||||
nFalseAlarms = nFalseAlarmsForThisTopN(ithThres);
|
||||
nHits = nHitsForThisTopN(ithThres);
|
||||
nMisses = nMissesForThisTopN(ithThres);
|
||||
|
||||
nTotalTestPlaces = nCorrectRejections + nFalseAlarms + nHits + nMisses;
|
||||
|
||||
Precision = nHits / (nHits + nFalseAlarms);
|
||||
Recall = nHits / (nHits + nMisses);
|
||||
Acc = (nHits + nCorrectRejections)/nTotalTestPlaces;
|
||||
|
||||
Precisions = [Precisions; Precision];
|
||||
Recalls = [Recalls; Recall];
|
||||
Accuracies = [Accuracies; Acc];
|
||||
end
|
||||
|
||||
num_points = length(Precisions);
|
||||
Precisions(1) = 1;
|
||||
AUC = 0;
|
||||
for ith = 1:num_points-1
|
||||
small_area = 1/2 * (Precisions(ith) + Precisions(ith+1)) * (Recalls(ith+1)-Recalls(ith));
|
||||
AUC = AUC + small_area;
|
||||
end
|
||||
AUCs(ithSeq) = AUC;
|
||||
|
||||
all_Precisions{ithSeq} = Precisions;
|
||||
all_Recalls{ithSeq} = Recalls;
|
||||
|
||||
% draw
|
||||
figure(FigIdx);
|
||||
set(gcf, 'Position', [10 10 800 500]);
|
||||
|
||||
fontsize = 10;
|
||||
p = plot(Recalls, Precisions, 'LineWidth', line_width); % commonly x axis is recall
|
||||
title(title_str, 'FontSize', fontsize);
|
||||
xlabel('Recall', 'FontSize', fontsize); ylabel('Precision', 'FontSize', fontsize);
|
||||
set(gca, 'FontSize', fontsize+5)
|
||||
xticks([0 0.2 0.4 0.6 0.8 1.0])
|
||||
xticklabels({'0','0.2','0.4','0.6','0.8','1'})
|
||||
yticks([0 0.2 0.4 0.6 0.8 1.0])
|
||||
yticklabels({'0','0.2','0.4','0.6','0.8','1'})
|
||||
|
||||
p(1).Color = LineColors(ithSeq, :);
|
||||
p(1).MarkerEdgeColor = LineColors(ithSeq, :);
|
||||
% axis equal;
|
||||
xlim([0, 1]); ylim([0,1]);
|
||||
grid on; grid minor;
|
||||
hold on;
|
||||
|
||||
end
|
||||
|
||||
lgd = legend(SequenceNames, 'Location', 'best');
|
||||
lgd.FontSize = fontsize + 3;
|
||||
lgd.FontWeight = 'bold';
|
||||
|
||||
grid minor;
|
||||
|
||||
name = 'prcurve';
|
||||
print('-bestfit', name,'-dpdf')
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,102 +0,0 @@
|
|||
function [ img ] = Ptcloud2ScanContext( ptcloud, num_sector, num_ring, max_range )
|
||||
|
||||
%% Preprocessing
|
||||
|
||||
% Downsampling for fast search
|
||||
gridStep = 0.5; % 0.5m cubic grid downsampling is applied in the paper.
|
||||
ptcloud = pcdownsample(ptcloud, 'gridAverage', gridStep);
|
||||
|
||||
% point cloud information
|
||||
num_points = ptcloud.Count;
|
||||
gap = max_range / num_ring;
|
||||
angle_one_sector = 360/num_sector;
|
||||
|
||||
|
||||
%% vacant bins
|
||||
cell_bins = cell(num_ring, num_sector);
|
||||
cell_bin_counter = ones(num_ring, num_sector);
|
||||
|
||||
enough_large = 500; % for fast and constant time save, We contain maximum 500 points per each bin.
|
||||
enough_small = -10000;
|
||||
for ith_ring = 1:num_ring
|
||||
for ith_sector = 1:num_sector
|
||||
bin = enough_small * ones(enough_large, 3);
|
||||
cell_bins{ith_ring, ith_sector} = bin;
|
||||
end
|
||||
end
|
||||
|
||||
|
||||
%% Save a point to the corresponding bin
|
||||
for ith_point =1:num_points
|
||||
|
||||
% Point information
|
||||
ith_point_xyz = ptcloud.Location(ith_point,:);
|
||||
ith_point_r = sqrt(ith_point_xyz(1)^2 + ith_point_xyz(2)^2);
|
||||
ith_point_theta = XY2Theta(ith_point_xyz(1), ith_point_xyz(2)); % degree
|
||||
|
||||
% Find the corresponding ring index
|
||||
tmp_ring_index = floor(ith_point_r/gap);
|
||||
if(tmp_ring_index >= num_ring)
|
||||
ring_index = num_ring;
|
||||
else
|
||||
ring_index = tmp_ring_index + 1;
|
||||
end
|
||||
|
||||
% Find the corresponding sector index
|
||||
tmp_sector_index = ceil(ith_point_theta/angle_one_sector);
|
||||
if(tmp_sector_index == 0)
|
||||
sector_index = 1;
|
||||
elseif(tmp_sector_index > num_sector || tmp_sector_index < 1)
|
||||
sector_index = num_sector;
|
||||
else
|
||||
sector_index = tmp_sector_index;
|
||||
end
|
||||
|
||||
% Assign point to the corresponding bin cell
|
||||
try
|
||||
corresponding_counter = cell_bin_counter(ring_index, sector_index); % 1D real value.
|
||||
catch
|
||||
continue;
|
||||
end
|
||||
cell_bins{ring_index, sector_index}(corresponding_counter, :) = ith_point_xyz;
|
||||
cell_bin_counter(ring_index, sector_index) = cell_bin_counter(ring_index, sector_index) + 1; % increase count 1
|
||||
|
||||
end
|
||||
|
||||
|
||||
%% bin to image format (2D matrix)
|
||||
img = zeros(num_ring, num_sector);
|
||||
|
||||
min_num_thres = 5; % a bin with few points, we consider it is noise.
|
||||
|
||||
% Find maximum Z value of each bin and Save into img
|
||||
for ith_ring = 1:num_ring
|
||||
for ith_sector = 1:num_sector
|
||||
value_of_the_bin = 0;
|
||||
points_in_bin_ij = cell_bins{ith_ring, ith_sector};
|
||||
|
||||
if( IsBinHaveMoreThanMinimumPoints(points_in_bin_ij, min_num_thres, enough_small) )
|
||||
value_of_the_bin = max(points_in_bin_ij(:, 3));
|
||||
else
|
||||
value_of_the_bin = 0;
|
||||
end
|
||||
|
||||
img(ith_ring, ith_sector) = value_of_the_bin;
|
||||
end
|
||||
end
|
||||
|
||||
|
||||
end % end of the main function
|
||||
|
||||
|
||||
function bool = IsBinHaveMoreThanMinimumPoints(mat, minimum_thres, enough_small)
|
||||
|
||||
min_thres_point = mat(minimum_thres, :);
|
||||
|
||||
if( isequal(min_thres_point, [ enough_small, enough_small, enough_small]) )
|
||||
bool = 0;
|
||||
else
|
||||
bool = 1;
|
||||
end
|
||||
|
||||
end
|
||||
|
|
@ -1,17 +0,0 @@
|
|||
function [ theta ] = XY2Theta( x, y )
|
||||
|
||||
if (x >= 0 && y >= 0)
|
||||
theta = 180/pi * atan(y/x);
|
||||
end
|
||||
if (x < 0 && y >= 0)
|
||||
theta = 180 - ((180/pi) * atan(y/(-x)));
|
||||
end
|
||||
if (x < 0 && y < 0)
|
||||
theta = 180 + ((180/pi) * atan(y/x));
|
||||
end
|
||||
if ( x >= 0 && y < 0)
|
||||
theta = 360 - ((180/pi) * atan((-y)/x));
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,121 +0,0 @@
|
|||
function [x,y,utmzone] = deg2utm(Lat,Lon)
|
||||
% -------------------------------------------------------------------------
|
||||
% [x,y,utmzone] = deg2utm(Lat,Lon)
|
||||
%
|
||||
% Description: Function to convert lat/lon vectors into UTM coordinates (WGS84).
|
||||
% Some code has been extracted from UTM.m function by Gabriel Ruiz Martinez.
|
||||
%
|
||||
% Inputs:
|
||||
% Lat: Latitude vector. Degrees. +ddd.ddddd WGS84
|
||||
% Lon: Longitude vector. Degrees. +ddd.ddddd WGS84
|
||||
%
|
||||
% Outputs:
|
||||
% x, y , utmzone. See example
|
||||
%
|
||||
% Example 1:
|
||||
% Lat=[40.3154333; 46.283900; 37.577833; 28.645650; 38.855550; 25.061783];
|
||||
% Lon=[-3.4857166; 7.8012333; -119.95525; -17.759533; -94.7990166; 121.640266];
|
||||
% [x,y,utmzone] = deg2utm(Lat,Lon);
|
||||
% fprintf('%7.0f ',x)
|
||||
% 458731 407653 239027 230253 343898 362850
|
||||
% fprintf('%7.0f ',y)
|
||||
% 4462881 5126290 4163083 3171843 4302285 2772478
|
||||
% utmzone =
|
||||
% 30 T
|
||||
% 32 T
|
||||
% 11 S
|
||||
% 28 R
|
||||
% 15 S
|
||||
% 51 R
|
||||
%
|
||||
% Example 2: If you have Lat/Lon coordinates in Degrees, Minutes and Seconds
|
||||
% LatDMS=[40 18 55.56; 46 17 2.04];
|
||||
% LonDMS=[-3 29 8.58; 7 48 4.44];
|
||||
% Lat=dms2deg(mat2dms(LatDMS)); %convert into degrees
|
||||
% Lon=dms2deg(mat2dms(LonDMS)); %convert into degrees
|
||||
% [x,y,utmzone] = deg2utm(Lat,Lon)
|
||||
%
|
||||
% Author:
|
||||
% Rafael Palacios
|
||||
% Universidad Pontificia Comillas
|
||||
% Madrid, Spain
|
||||
% Version: Apr/06, Jun/06, Aug/06, Aug/06
|
||||
% Aug/06: fixed a problem (found by Rodolphe Dewarrat) related to southern
|
||||
% hemisphere coordinates.
|
||||
% Aug/06: corrected m-Lint warnings
|
||||
%-------------------------------------------------------------------------
|
||||
% Argument checking
|
||||
%
|
||||
error(nargchk(2, 2, nargin)); %2 arguments required
|
||||
n1=length(Lat);
|
||||
n2=length(Lon);
|
||||
if (n1~=n2)
|
||||
error('Lat and Lon vectors should have the same length');
|
||||
end
|
||||
% Memory pre-allocation
|
||||
%
|
||||
x=zeros(n1,1);
|
||||
y=zeros(n1,1);
|
||||
utmzone(n1,:)='60 X';
|
||||
% Main Loop
|
||||
%
|
||||
for i=1:n1
|
||||
la=Lat(i);
|
||||
lo=Lon(i);
|
||||
sa = 6378137.000000 ; sb = 6356752.314245;
|
||||
|
||||
%e = ( ( ( sa ^ 2 ) - ( sb ^ 2 ) ) ^ 0.5 ) / sa;
|
||||
e2 = ( ( ( sa ^ 2 ) - ( sb ^ 2 ) ) ^ 0.5 ) / sb;
|
||||
e2cuadrada = e2 ^ 2;
|
||||
c = ( sa ^ 2 ) / sb;
|
||||
%alpha = ( sa - sb ) / sa; %f
|
||||
%ablandamiento = 1 / alpha; % 1/f
|
||||
lat = la * ( pi / 180 );
|
||||
lon = lo * ( pi / 180 );
|
||||
Huso = fix( ( lo / 6 ) + 31);
|
||||
S = ( ( Huso * 6 ) - 183 );
|
||||
deltaS = lon - ( S * ( pi / 180 ) );
|
||||
if (la<-72), Letra='C';
|
||||
elseif (la<-64), Letra='D';
|
||||
elseif (la<-56), Letra='E';
|
||||
elseif (la<-48), Letra='F';
|
||||
elseif (la<-40), Letra='G';
|
||||
elseif (la<-32), Letra='H';
|
||||
elseif (la<-24), Letra='J';
|
||||
elseif (la<-16), Letra='K';
|
||||
elseif (la<-8), Letra='L';
|
||||
elseif (la<0), Letra='M';
|
||||
elseif (la<8), Letra='N';
|
||||
elseif (la<16), Letra='P';
|
||||
elseif (la<24), Letra='Q';
|
||||
elseif (la<32), Letra='R';
|
||||
elseif (la<40), Letra='S';
|
||||
elseif (la<48), Letra='T';
|
||||
elseif (la<56), Letra='U';
|
||||
elseif (la<64), Letra='V';
|
||||
elseif (la<72), Letra='W';
|
||||
else Letra='X';
|
||||
end
|
||||
a = cos(lat) * sin(deltaS);
|
||||
epsilon = 0.5 * log( ( 1 + a) / ( 1 - a ) );
|
||||
nu = atan( tan(lat) / cos(deltaS) ) - lat;
|
||||
v = ( c / ( ( 1 + ( e2cuadrada * ( cos(lat) ) ^ 2 ) ) ) ^ 0.5 ) * 0.9996;
|
||||
ta = ( e2cuadrada / 2 ) * epsilon ^ 2 * ( cos(lat) ) ^ 2;
|
||||
a1 = sin( 2 * lat );
|
||||
a2 = a1 * ( cos(lat) ) ^ 2;
|
||||
j2 = lat + ( a1 / 2 );
|
||||
j4 = ( ( 3 * j2 ) + a2 ) / 4;
|
||||
j6 = ( ( 5 * j4 ) + ( a2 * ( cos(lat) ) ^ 2) ) / 3;
|
||||
alfa = ( 3 / 4 ) * e2cuadrada;
|
||||
beta = ( 5 / 3 ) * alfa ^ 2;
|
||||
gama = ( 35 / 27 ) * alfa ^ 3;
|
||||
Bm = 0.9996 * c * ( lat - alfa * j2 + beta * j4 - gama * j6 );
|
||||
xx = epsilon * v * ( 1 + ( ta / 3 ) ) + 500000;
|
||||
yy = nu * v * ( 1 + ta ) + Bm;
|
||||
if (yy<0)
|
||||
yy=9999999+yy;
|
||||
end
|
||||
x(i)=xx;
|
||||
y(i)=yy;
|
||||
utmzone(i,:)=sprintf('%02d %c',Huso,Letra);
|
||||
end
|
||||
|
|
@ -1,3 +0,0 @@
|
|||
function dist = dist_btn_pose(pose1, pose2)
|
||||
dist = sqrt( (pose1(1) - pose2(1))^2 + (pose1(2) - pose2(2))^2);
|
||||
end
|
||||
|
|
@ -1,28 +0,0 @@
|
|||
function [ nearest_idx, min_dist ] = find_topk_from_candidates(query_img, query_idx, candidates, thres)
|
||||
|
||||
global num_node_enough_apart;
|
||||
global radar_imgs;
|
||||
|
||||
nearest_idx = 0;
|
||||
min_dist = inf; % initialization
|
||||
for ith_candidate = 1:length(candidates)
|
||||
candidate_node_idx = candidates(ith_candidate);
|
||||
candidate_img = radar_imgs{candidate_node_idx};
|
||||
|
||||
if( abs(query_idx - candidate_node_idx) < num_node_enough_apart)
|
||||
continue;
|
||||
end
|
||||
|
||||
distance_to_query = dist(query_img, candidate_img);
|
||||
if( distance_to_query > thres)
|
||||
continue;
|
||||
end
|
||||
|
||||
if( distance_to_query < min_dist)
|
||||
nearest_idx = candidate_node_idx;
|
||||
min_dist = distance_to_query;
|
||||
end
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,20 +0,0 @@
|
|||
function [is_revisit, min_dist] = isRevisitGlobalLoc(query_pose, db_poses, thres)
|
||||
|
||||
num_dbs = length(db_poses);
|
||||
|
||||
dists = zeros(1, num_dbs);
|
||||
for ii=1:num_dbs
|
||||
dist = norm(query_pose - db_poses(ii, :));
|
||||
dists(ii) = dist;
|
||||
end
|
||||
|
||||
if ( min(dists) < thres )
|
||||
is_revisit = 1;
|
||||
else
|
||||
is_revisit = 0;
|
||||
end
|
||||
|
||||
min_dist = min(dists);
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,23 +0,0 @@
|
|||
function [ revisitness ] = is_revisit(query_idx, query_pose, radar_poses, revisit_criteria, num_node_enough_apart)
|
||||
|
||||
num_db = size(radar_poses, 1);
|
||||
|
||||
revisitness = 0;
|
||||
for ii = 1:num_db
|
||||
|
||||
if( abs(query_idx - ii) < num_node_enough_apart)
|
||||
continue;
|
||||
end
|
||||
|
||||
pose = radar_poses(ii, :);
|
||||
|
||||
dist = dist_btn_pose(query_pose, pose);
|
||||
|
||||
if(dist < revisit_criteria)
|
||||
revisitness = 1;
|
||||
break;
|
||||
end
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,45 +0,0 @@
|
|||
function [scancontexts, ringkeys, poses] = loadData(down_shape, skip_data_frame)
|
||||
|
||||
%%
|
||||
global data_path;
|
||||
data_save_path = fullfile('data/');
|
||||
|
||||
%%
|
||||
% newly make
|
||||
if exist(data_save_path) == 0
|
||||
% make
|
||||
[scancontexts, ringkeys, poses] = makeExperience(data_path, down_shape, skip_data_frame);
|
||||
|
||||
% save
|
||||
mkdir(data_save_path);
|
||||
|
||||
filename = strcat(data_save_path, 'scancontexts', num2str(down_shape(1)), 'x', num2str(down_shape(2)), '.mat');
|
||||
save(filename, 'scancontexts');
|
||||
filename = strcat(data_save_path, 'ringkeys', num2str(down_shape(1)), 'x', num2str(down_shape(2)), '.mat');
|
||||
save(filename, 'ringkeys');
|
||||
filename = strcat(data_save_path, 'poses', num2str(down_shape(1)), 'x', num2str(down_shape(2)), '.mat');
|
||||
save(filename, 'poses');
|
||||
|
||||
% or load
|
||||
else
|
||||
filename = strcat(data_save_path, 'scancontexts', num2str(down_shape(1)), 'x', num2str(down_shape(2)), '.mat');
|
||||
load(filename);
|
||||
% fix
|
||||
for iii = 1:length(scancontexts)
|
||||
sc = double(scancontexts{iii});
|
||||
scancontexts{iii} = sc;
|
||||
end
|
||||
|
||||
filename = strcat(data_save_path, 'ringkeys', num2str(down_shape(1)), 'x', num2str(down_shape(2)), '.mat');
|
||||
load(filename);
|
||||
filename = strcat(data_save_path, 'poses', num2str(down_shape(1)), 'x', num2str(down_shape(2)), '.mat');
|
||||
load(filename);
|
||||
|
||||
disp('- successfully loaded.');
|
||||
end
|
||||
|
||||
%%
|
||||
disp(' ');
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,63 +0,0 @@
|
|||
function [scancontexts, ringkeys, xy_poses] = makeExperience(data_dir, shape, skip_data_frame)
|
||||
|
||||
%%
|
||||
num_rings = shape(1);
|
||||
num_sectors = shape(2);
|
||||
|
||||
%%
|
||||
lidar_data_dir = strcat(data_dir, 'velodyne/');
|
||||
data_names = osdir(lidar_data_dir);
|
||||
|
||||
%% gps to xyz
|
||||
gtpose = csvread(strcat(data_dir, '00.csv'));
|
||||
% gtpose_time = gtpose(:, 1);
|
||||
gtpose_xy = gtpose(:, [4,12]);
|
||||
|
||||
|
||||
%%
|
||||
num_data = length(data_names);
|
||||
num_data_save = floor(num_data/skip_data_frame) + 1;
|
||||
save_counter = 1;
|
||||
|
||||
scancontexts = cell(1, num_data_save);
|
||||
ringkeys = zeros(num_data_save, num_rings);
|
||||
xy_poses = zeros(num_data_save, 2);
|
||||
|
||||
for data_idx = 1:num_data
|
||||
|
||||
if(rem(data_idx, skip_data_frame) ~=0)
|
||||
continue;
|
||||
end
|
||||
|
||||
file_name = data_names{data_idx};
|
||||
data_time = str2double(file_name(1:end-4));
|
||||
data_path = strcat(lidar_data_dir, file_name);
|
||||
|
||||
% get
|
||||
ptcloud = readBin(data_path);
|
||||
sc = Ptcloud2ScanContext(ptcloud, shape(2), shape(1), 80); % up to 80 meter
|
||||
|
||||
rk = ringkey(sc);
|
||||
|
||||
% [nearest_time_gap, nearest_idx] = min(abs(repmat(data_time, length(gtpose_time), 1) - gtpose_time));
|
||||
xy_pose = gtpose_xy(data_idx, :);
|
||||
|
||||
% save
|
||||
scancontexts{save_counter} = sc;
|
||||
ringkeys(save_counter, :) = rk;
|
||||
xy_poses(save_counter, :) = xy_pose;
|
||||
save_counter = save_counter + 1;
|
||||
|
||||
% log
|
||||
if(rem(data_idx, 100) == 0)
|
||||
message = strcat(num2str(data_idx), " / ", num2str(num_data), " processed (skip: ", num2str(skip_data_frame), ")");
|
||||
disp(message);
|
||||
end
|
||||
end
|
||||
|
||||
scancontexts = scancontexts(1:save_counter-1);
|
||||
ringkeys = ringkeys(1:save_counter-1, :);
|
||||
xy_poses = xy_poses(1:save_counter-1, :);
|
||||
|
||||
|
||||
end
|
||||
|
|
@ -1,4 +0,0 @@
|
|||
function [files] = osdir(path)
|
||||
files = dir(path); files(1:2) = []; files = {files(:).name};
|
||||
end
|
||||
|
||||
|
|
@ -1,10 +0,0 @@
|
|||
function ptcloud = readBin(bin_path)
|
||||
|
||||
%% Read
|
||||
fid = fopen(bin_path, 'rb'); raw_data = fread(fid, [4 inf], 'single'); fclose(fid);
|
||||
points = raw_data(1:3,:)';
|
||||
points(:, 3) = points(:, 3) + 1.9; % z in car coord.
|
||||
|
||||
ptcloud = pointCloud(points);
|
||||
|
||||
end % end of function
|
||||
|
|
@ -1,29 +0,0 @@
|
|||
function [down_img] = resize_polar_img(varargin)
|
||||
|
||||
%%
|
||||
% arg 1: target image
|
||||
% arg 2: size of the downsized image; the number of [r, theta] for 200m, 360 deg
|
||||
% arg 3: interpolation type
|
||||
|
||||
%%
|
||||
if nargin == 1
|
||||
img = varargin{1};
|
||||
rescale_pixel = [40, 60];
|
||||
interpolation_method = 'box';
|
||||
end
|
||||
|
||||
if nargin == 2
|
||||
img = varargin{1};
|
||||
rescale_pixel = varargin{2};
|
||||
interpolation_method = 'box';
|
||||
end
|
||||
|
||||
if nargin == 3
|
||||
img = varargin{1};
|
||||
rescale_pixel = varargin{2};
|
||||
interpolation_method = varargin{3};
|
||||
end
|
||||
|
||||
down_img = imresize(img, rescale_pixel, 'method', interpolation_method);
|
||||
|
||||
end
|
||||
|
|
@ -1,13 +0,0 @@
|
|||
function [ ring_key ] = ringkey(sc)
|
||||
|
||||
num_rings = size(sc, 1);
|
||||
|
||||
ring_key = zeros(1, num_rings);
|
||||
for ith=1:num_rings
|
||||
ith_ring = sc(ith,:);
|
||||
% ring_key(ith) = mean(ith_ring);
|
||||
ring_key(ith) = nnz(ith_ring);
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
|
@ -1,41 +0,0 @@
|
|||
function [dist] = dist(sc1,sc2)
|
||||
|
||||
num_sectors = size(sc1, 2);
|
||||
|
||||
% repeate to move 1 columns
|
||||
sim_for_each_cols = zeros(1, num_sectors);
|
||||
|
||||
for i = 1:num_sectors
|
||||
%% Shift
|
||||
one_step = 1; % const
|
||||
sc1 = circshift(sc1, one_step, 2); % 2 means columne shift
|
||||
|
||||
%% compare
|
||||
sum_of_cos_sim = 0;
|
||||
num_col_engaged = 0;
|
||||
|
||||
for j = 1:num_sectors
|
||||
col_j_1 = sc1(:,j);
|
||||
col_j_2 = sc2(:,j);
|
||||
|
||||
if( ~any(col_j_1) || ~any(col_j_2))
|
||||
continue;
|
||||
end
|
||||
|
||||
% calc sim
|
||||
cos_similarity = dot(col_j_1, col_j_2) / (norm(col_j_1)*norm(col_j_2));
|
||||
sum_of_cos_sim = sum_of_cos_sim + cos_similarity;
|
||||
|
||||
num_col_engaged = num_col_engaged +1;
|
||||
end
|
||||
|
||||
% devided by num_col_engaged: So, even if there are many columns that are excluded from the calculation, we can get high scores if other columns are well fit.
|
||||
sim_for_each_cols(i) = sum_of_cos_sim/num_col_engaged;
|
||||
|
||||
end
|
||||
|
||||
sim = max(sim_for_each_cols);
|
||||
|
||||
dist = 1 - sim;
|
||||
|
||||
end
|
||||
|
|
@ -1,23 +0,0 @@
|
|||
# Radar Scan Context
|
||||
|
||||
- Scan Context also works for the radar data (i.e., Navtech radar)
|
||||
- Radar Scan Context was introduced in the [MulRan dataset paper](https://irap.kaist.ac.kr/publications/gskim-2020-icra.pdf)
|
||||
- This directory contains the evaluation code, for [radar place recognition](https://sites.google.com/view/mulran-pr/radar-place-recognition), used in the MulRan paper.
|
||||
- if you use the dataset or our method, please refer the paper:
|
||||
```
|
||||
@INPROCEEDINGS { gskim-2020-icra,
|
||||
AUTHOR = { Giseop Kim, Yeong Sang Park, Younghun Cho, Jinyong Jeong, Ayoung Kim },
|
||||
TITLE = { MulRan: Multimodal Range Dataset for Urban Place Recognition },
|
||||
BOOKTITLE = { Proceedings of the IEEE International Conference on Robotics and Automation (ICRA) },
|
||||
YEAR = { 2020 },
|
||||
MONTH = { May },
|
||||
ADDRESS = { Paris }
|
||||
}
|
||||
```
|
||||
|
||||
- More information about the radar data, please refer [MulRan](https://sites.google.com/view/mulran-pr/home) or Oxford Radar RobotCar dataset
|
||||
|
||||
## How to use
|
||||
- 1. write your own MulRan dataset path in the main.m file
|
||||
- 2. run main.m (then some data and evaluation files will be generated)
|
||||
- 3. run prcurve_drawer.m
|
||||
|
|
@ -1,261 +0,0 @@
|
|||
% function lineStyles = linspecer(N)
|
||||
% This function creates an Nx3 array of N [R B G] colors
|
||||
% These can be used to plot lots of lines with distinguishable and nice
|
||||
% looking colors.
|
||||
%
|
||||
% lineStyles = linspecer(N); makes N colors for you to use: lineStyles(ii,:)
|
||||
%
|
||||
% colormap(linspecer); set your colormap to have easily distinguishable
|
||||
% colors and a pleasing aesthetic
|
||||
%
|
||||
% lineStyles = linspecer(N,'qualitative'); forces the colors to all be distinguishable (up to 12)
|
||||
% lineStyles = linspecer(N,'sequential'); forces the colors to vary along a spectrum
|
||||
%
|
||||
% % Examples demonstrating the colors.
|
||||
%
|
||||
% LINE COLORS
|
||||
% N=6;
|
||||
% X = linspace(0,pi*3,1000);
|
||||
% Y = bsxfun(@(x,n)sin(x+2*n*pi/N), X.', 1:N);
|
||||
% C = linspecer(N);
|
||||
% axes('NextPlot','replacechildren', 'ColorOrder',C);
|
||||
% plot(X,Y,'linewidth',5)
|
||||
% ylim([-1.1 1.1]);
|
||||
%
|
||||
% SIMPLER LINE COLOR EXAMPLE
|
||||
% N = 6; X = linspace(0,pi*3,1000);
|
||||
% C = linspecer(N)
|
||||
% hold off;
|
||||
% for ii=1:N
|
||||
% Y = sin(X+2*ii*pi/N);
|
||||
% plot(X,Y,'color',C(ii,:),'linewidth',3);
|
||||
% hold on;
|
||||
% end
|
||||
%
|
||||
% COLORMAP EXAMPLE
|
||||
% A = rand(15);
|
||||
% figure; imagesc(A); % default colormap
|
||||
% figure; imagesc(A); colormap(linspecer); % linspecer colormap
|
||||
%
|
||||
% See also NDHIST, NHIST, PLOT, COLORMAP, 43700-cubehelix-colormaps
|
||||
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
|
||||
% by Jonathan Lansey, March 2009-2013 – Lansey at gmail.com %
|
||||
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
|
||||
%
|
||||
%% credits and where the function came from
|
||||
% The colors are largely taken from:
|
||||
% http://colorbrewer2.org and Cynthia Brewer, Mark Harrower and The Pennsylvania State University
|
||||
%
|
||||
%
|
||||
% She studied this from a phsychometric perspective and crafted the colors
|
||||
% beautifully.
|
||||
%
|
||||
% I made choices from the many there to decide the nicest once for plotting
|
||||
% lines in Matlab. I also made a small change to one of the colors I
|
||||
% thought was a bit too bright. In addition some interpolation is going on
|
||||
% for the sequential line styles.
|
||||
%
|
||||
%
|
||||
%%
|
||||
|
||||
function lineStyles=linspecer(N,varargin)
|
||||
|
||||
if nargin==0 % return a colormap
|
||||
lineStyles = linspecer(128);
|
||||
return;
|
||||
end
|
||||
|
||||
if ischar(N)
|
||||
lineStyles = linspecer(128,N);
|
||||
return;
|
||||
end
|
||||
|
||||
if N<=0 % its empty, nothing else to do here
|
||||
lineStyles=[];
|
||||
return;
|
||||
end
|
||||
|
||||
% interperet varagin
|
||||
qualFlag = 0;
|
||||
colorblindFlag = 0;
|
||||
|
||||
if ~isempty(varargin)>0 % you set a parameter?
|
||||
switch lower(varargin{1})
|
||||
case {'qualitative','qua'}
|
||||
if N>12 % go home, you just can't get this.
|
||||
warning('qualitiative is not possible for greater than 12 items, please reconsider');
|
||||
else
|
||||
if N>9
|
||||
warning(['Default may be nicer for ' num2str(N) ' for clearer colors use: whitebg(''black''); ']);
|
||||
end
|
||||
end
|
||||
qualFlag = 1;
|
||||
case {'sequential','seq'}
|
||||
lineStyles = colorm(N);
|
||||
return;
|
||||
case {'white','whitefade'}
|
||||
lineStyles = whiteFade(N);return;
|
||||
case 'red'
|
||||
lineStyles = whiteFade(N,'red');return;
|
||||
case 'blue'
|
||||
lineStyles = whiteFade(N,'blue');return;
|
||||
case 'green'
|
||||
lineStyles = whiteFade(N,'green');return;
|
||||
case {'gray','grey'}
|
||||
lineStyles = whiteFade(N,'gray');return;
|
||||
case {'colorblind'}
|
||||
colorblindFlag = 1;
|
||||
otherwise
|
||||
warning(['parameter ''' varargin{1} ''' not recognized']);
|
||||
end
|
||||
end
|
||||
% *.95
|
||||
% predefine some colormaps
|
||||
set3 = colorBrew2mat({[141, 211, 199];[ 255, 237, 111];[ 190, 186, 218];[ 251, 128, 114];[ 128, 177, 211];[ 253, 180, 98];[ 179, 222, 105];[ 188, 128, 189];[ 217, 217, 217];[ 204, 235, 197];[ 252, 205, 229];[ 255, 255, 179]}');
|
||||
set1JL = brighten(colorBrew2mat({[228, 26, 28];[ 55, 126, 184]; [ 77, 175, 74];[ 255, 127, 0];[ 255, 237, 111]*.85;[ 166, 86, 40];[ 247, 129, 191];[ 153, 153, 153];[ 152, 78, 163]}'));
|
||||
set1 = brighten(colorBrew2mat({[ 55, 126, 184]*.85;[228, 26, 28];[ 77, 175, 74];[ 255, 127, 0];[ 152, 78, 163]}),.8);
|
||||
|
||||
% colorblindSet = {[215,25,28];[253,174,97];[171,217,233];[44,123,182]};
|
||||
colorblindSet = {[215,25,28];[253,174,97];[171,217,233]*.8;[44,123,182]*.8};
|
||||
|
||||
set3 = dim(set3,.93);
|
||||
|
||||
if colorblindFlag
|
||||
switch N
|
||||
% sorry about this line folks. kind of legacy here because I used to
|
||||
% use individual 1x3 cells instead of nx3 arrays
|
||||
case 4
|
||||
lineStyles = colorBrew2mat(colorblindSet);
|
||||
otherwise
|
||||
colorblindFlag = false;
|
||||
warning('sorry unsupported colorblind set for this number, using regular types');
|
||||
end
|
||||
end
|
||||
if ~colorblindFlag
|
||||
switch N
|
||||
case 1
|
||||
lineStyles = { [ 55, 126, 184]/255};
|
||||
case {2, 3, 4, 5 }
|
||||
lineStyles = set1(1:N);
|
||||
case {6 , 7, 8, 9}
|
||||
lineStyles = set1JL(1:N)';
|
||||
case {10, 11, 12}
|
||||
if qualFlag % force qualitative graphs
|
||||
lineStyles = set3(1:N)';
|
||||
else % 10 is a good number to start with the sequential ones.
|
||||
lineStyles = cmap2linspecer(colorm(N));
|
||||
end
|
||||
otherwise % any old case where I need a quick job done.
|
||||
lineStyles = cmap2linspecer(colorm(N));
|
||||
end
|
||||
end
|
||||
lineStyles = cell2mat(lineStyles);
|
||||
|
||||
end
|
||||
|
||||
% extra functions
|
||||
function varIn = colorBrew2mat(varIn)
|
||||
for ii=1:length(varIn) % just divide by 255
|
||||
varIn{ii}=varIn{ii}/255;
|
||||
end
|
||||
end
|
||||
|
||||
function varIn = brighten(varIn,varargin) % increase the brightness
|
||||
|
||||
if isempty(varargin),
|
||||
frac = .9;
|
||||
else
|
||||
frac = varargin{1};
|
||||
end
|
||||
|
||||
for ii=1:length(varIn)
|
||||
varIn{ii}=varIn{ii}*frac+(1-frac);
|
||||
end
|
||||
end
|
||||
|
||||
function varIn = dim(varIn,f)
|
||||
for ii=1:length(varIn)
|
||||
varIn{ii} = f*varIn{ii};
|
||||
end
|
||||
end
|
||||
|
||||
function vOut = cmap2linspecer(vIn) % changes the format from a double array to a cell array with the right format
|
||||
vOut = cell(size(vIn,1),1);
|
||||
for ii=1:size(vIn,1)
|
||||
vOut{ii} = vIn(ii,:);
|
||||
end
|
||||
end
|
||||
%%
|
||||
% colorm returns a colormap which is really good for creating informative
|
||||
% heatmap style figures.
|
||||
% No particular color stands out and it doesn't do too badly for colorblind people either.
|
||||
% It works by interpolating the data from the
|
||||
% 'spectral' setting on http://colorbrewer2.org/ set to 11 colors
|
||||
% It is modified a little to make the brightest yellow a little less bright.
|
||||
function cmap = colorm(varargin)
|
||||
n = 100;
|
||||
if ~isempty(varargin)
|
||||
n = varargin{1};
|
||||
end
|
||||
|
||||
if n==1
|
||||
cmap = [0.2005 0.5593 0.7380];
|
||||
return;
|
||||
end
|
||||
if n==2
|
||||
cmap = [0.2005 0.5593 0.7380;
|
||||
0.9684 0.4799 0.2723];
|
||||
return;
|
||||
end
|
||||
|
||||
frac=.95; % Slight modification from colorbrewer here to make the yellows in the center just a bit darker
|
||||
cmapp = [158, 1, 66; 213, 62, 79; 244, 109, 67; 253, 174, 97; 254, 224, 139; 255*frac, 255*frac, 191*frac; 230, 245, 152; 171, 221, 164; 102, 194, 165; 50, 136, 189; 94, 79, 162];
|
||||
x = linspace(1,n,size(cmapp,1));
|
||||
xi = 1:n;
|
||||
cmap = zeros(n,3);
|
||||
for ii=1:3
|
||||
cmap(:,ii) = pchip(x,cmapp(:,ii),xi);
|
||||
end
|
||||
cmap = flipud(cmap/255);
|
||||
end
|
||||
|
||||
function cmap = whiteFade(varargin)
|
||||
n = 100;
|
||||
if nargin>0
|
||||
n = varargin{1};
|
||||
end
|
||||
|
||||
thisColor = 'blue';
|
||||
|
||||
if nargin>1
|
||||
thisColor = varargin{2};
|
||||
end
|
||||
switch thisColor
|
||||
case {'gray','grey'}
|
||||
cmapp = [255,255,255;240,240,240;217,217,217;189,189,189;150,150,150;115,115,115;82,82,82;37,37,37;0,0,0];
|
||||
case 'green'
|
||||
cmapp = [247,252,245;229,245,224;199,233,192;161,217,155;116,196,118;65,171,93;35,139,69;0,109,44;0,68,27];
|
||||
case 'blue'
|
||||
cmapp = [247,251,255;222,235,247;198,219,239;158,202,225;107,174,214;66,146,198;33,113,181;8,81,156;8,48,107];
|
||||
case 'red'
|
||||
cmapp = [255,245,240;254,224,210;252,187,161;252,146,114;251,106,74;239,59,44;203,24,29;165,15,21;103,0,13];
|
||||
otherwise
|
||||
warning(['sorry your color argument ' thisColor ' was not recognized']);
|
||||
end
|
||||
|
||||
cmap = interpomap(n,cmapp);
|
||||
end
|
||||
|
||||
% Eat a approximate colormap, then interpolate the rest of it up.
|
||||
function cmap = interpomap(n,cmapp)
|
||||
x = linspace(1,n,size(cmapp,1));
|
||||
xi = 1:n;
|
||||
cmap = zeros(n,3);
|
||||
for ii=1:3
|
||||
cmap(:,ii) = pchip(x,cmapp(:,ii),xi);
|
||||
end
|
||||
cmap = (cmap/255); % flipud??
|
||||
end
|
||||
|
||||
|
||||
|
||||
|
|
@ -1,162 +0,0 @@
|
|||
clear; clc;
|
||||
|
||||
addpath(genpath('src'));
|
||||
addpath(genpath('data'));
|
||||
|
||||
%% data preparation
|
||||
global data_path;
|
||||
% data_path = '/your/mulran/sequence/dir/Riverside02/';
|
||||
data_path = '/media/user/My Passport/data/MulRan_eval/Riverside_2_20190816/20190816/';
|
||||
% ### NOTE: Use this sequence directory structure
|
||||
% example:
|
||||
% /your/MulRan/sequence/dir/Riverside02/
|
||||
% L sensor_data/
|
||||
% L radar/
|
||||
% L polar/
|
||||
% L {unix_times}.png
|
||||
% L global_pose.csv
|
||||
|
||||
down_shape = [40, 120];
|
||||
[data_scancontexts, data_ringkeys, data_poses] = loadData(down_shape);
|
||||
|
||||
|
||||
%% main - global recognizer
|
||||
revisit_criteria = 5; % in meter (recommend test for 5, 10, 20 meters)
|
||||
keyframe_gap = 1; % for_fast_eval (if 1, no skip)
|
||||
|
||||
global num_candidates; num_candidates = 5;
|
||||
% NOTE about num_candidates
|
||||
% - 50 was used in the MulRan paper
|
||||
% - But we found, interestingly, using less keys showed similar
|
||||
% performance - also we can save the computation time of course.
|
||||
% - That means our ring key has good disriminative power.
|
||||
|
||||
global num_node_enough_apart; num_node_enough_apart = 50;
|
||||
|
||||
% policy (top N)
|
||||
num_top_n = 25;
|
||||
top_n = linspace(1, num_top_n, num_top_n);
|
||||
|
||||
% Entropy thresholds
|
||||
middle_thres = 0.01;
|
||||
thresholds1 = linspace(0, middle_thres, 50);
|
||||
thresholds2 = linspace(middle_thres, 1, 50);
|
||||
thresholds = [thresholds1, thresholds2];
|
||||
num_thresholds = length(thresholds);
|
||||
|
||||
% Main variables to store the result for drawing PR curve
|
||||
num_hits = zeros(num_top_n, num_thresholds);
|
||||
num_false_alarms = zeros(num_top_n, num_thresholds);
|
||||
num_correct_rejections = zeros(num_top_n, num_thresholds);
|
||||
num_misses = zeros(num_top_n, num_thresholds);
|
||||
|
||||
% main
|
||||
loop_log = [];
|
||||
|
||||
exp_poses = [];
|
||||
exp_ringkeys = [];
|
||||
exp_scancontexts = {};
|
||||
|
||||
num_queries = length(data_poses);
|
||||
for query_idx = 1:num_queries - 1
|
||||
|
||||
% save to (online) DB
|
||||
query_sc = data_scancontexts{query_idx};
|
||||
query_rk = data_ringkeys(query_idx, :);
|
||||
query_pose = data_poses(query_idx,:);
|
||||
|
||||
exp_scancontexts{end+1} = query_sc;
|
||||
exp_poses = [exp_poses; query_pose];
|
||||
exp_ringkeys = [exp_ringkeys; query_rk];
|
||||
|
||||
if(rem(query_idx, keyframe_gap) ~= 0)
|
||||
continue;
|
||||
end
|
||||
|
||||
if( length(exp_scancontexts) < num_node_enough_apart )
|
||||
continue;
|
||||
end
|
||||
|
||||
tree = createns(exp_ringkeys(1:end-(num_node_enough_apart-1), :), 'NSMethod', 'kdtree'); % Create object to use in k-nearest neighbor search
|
||||
|
||||
% revisitness
|
||||
[revisitness, how_far_apart] = isRevisitGlobalLoc(query_pose, exp_poses(1:end-(num_node_enough_apart-1), :), revisit_criteria);
|
||||
|
||||
% find candidates
|
||||
candidates = knnsearch(tree, query_rk, 'K', num_candidates);
|
||||
|
||||
% find the nearest (top 1) via pairwise comparison
|
||||
nearest_idx = 0;
|
||||
min_dist = inf; % initialization
|
||||
for ith_candidate = 1:length(candidates)
|
||||
candidate_node_idx = candidates(ith_candidate);
|
||||
candidate_img = exp_scancontexts{candidate_node_idx};
|
||||
|
||||
% if( abs(query_idx - candidate_node_idx) < num_node_enough_apart)
|
||||
% continue;
|
||||
% end
|
||||
|
||||
distance_to_query = sc_dist(query_sc, candidate_img);
|
||||
|
||||
if( distance_to_query < min_dist)
|
||||
nearest_idx = candidate_node_idx;
|
||||
min_dist = distance_to_query;
|
||||
end
|
||||
end
|
||||
|
||||
% prcurve analysis
|
||||
for topk = 1:num_top_n
|
||||
for thres_idx = 1:num_thresholds
|
||||
threshold = thresholds(thres_idx);
|
||||
|
||||
reject = 0;
|
||||
if( min_dist > threshold)
|
||||
reject = 1;
|
||||
end
|
||||
|
||||
if(reject == 1)
|
||||
if(revisitness == 0)
|
||||
% TN: Correct Rejection
|
||||
num_correct_rejections(topk, thres_idx) = num_correct_rejections(topk, thres_idx) + 1;
|
||||
else
|
||||
% FN: MISS
|
||||
num_misses(topk, thres_idx) = num_misses(topk, thres_idx) + 1;
|
||||
end
|
||||
else
|
||||
% if under the theshold, it is considered seen.
|
||||
% and then check the correctness
|
||||
if( dist_btn_pose(query_pose, exp_poses(nearest_idx, :)) < revisit_criteria)
|
||||
% TP: Hit
|
||||
num_hits(topk, thres_idx) = num_hits(topk, thres_idx) + 1;
|
||||
else
|
||||
% FP: False Alarm
|
||||
num_false_alarms(topk, thres_idx) = num_false_alarms(topk, thres_idx) + 1;
|
||||
end
|
||||
end
|
||||
|
||||
end
|
||||
end
|
||||
|
||||
if( rem(query_idx, 100) == 0)
|
||||
disp( strcat(num2str(query_idx/num_queries * 100), ' % processed') );
|
||||
end
|
||||
|
||||
end
|
||||
|
||||
|
||||
%% save the log
|
||||
savePath = strcat("pr_result/within ", num2str(revisit_criteria), "m/");
|
||||
if((~7==exist(savePath,'dir')))
|
||||
mkdir(savePath);
|
||||
end
|
||||
save(strcat(savePath, 'nCorrectRejections.mat'), 'num_correct_rejections');
|
||||
save(strcat(savePath, 'nMisses.mat'), 'num_misses');
|
||||
save(strcat(savePath, 'nHits.mat'), 'num_hits');
|
||||
save(strcat(savePath, 'nFalseAlarms.mat'), 'num_false_alarms');
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
Some files were not shown because too many files have changed in this diff Show More
Loading…
Reference in New Issue